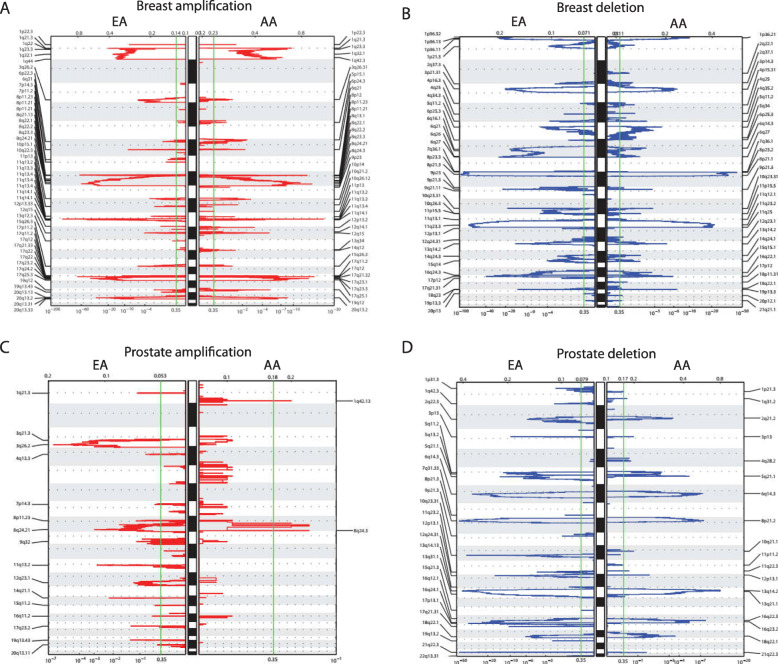
Fig. 1.
Autosomal SCNAs identified by Gistic2. a Breast tumor amplification SCNAs in European American (EA) and African American (AA) breast tumors. b Breast tumor deletion SCNAs in EAs and AAs. c Prostate tumor amplification SCNAs in EAs and AAs. d Prostate tumor deletion SCNAs in EAs and AAs. The autosomes are arranged in chromosomal order from top to bottom, and the horizontal dotted lines indicate the centromere for each chromosome. The cytobands are plotted on the left and right of each panel only for significant SCNAs. The bottom axis shows the false discovery rate (FDR) for SCNA detection, and the green line indicates the default significance threshold (FDR < 0.35) used by Gistic2 to identify SCNAs