Figure 1.
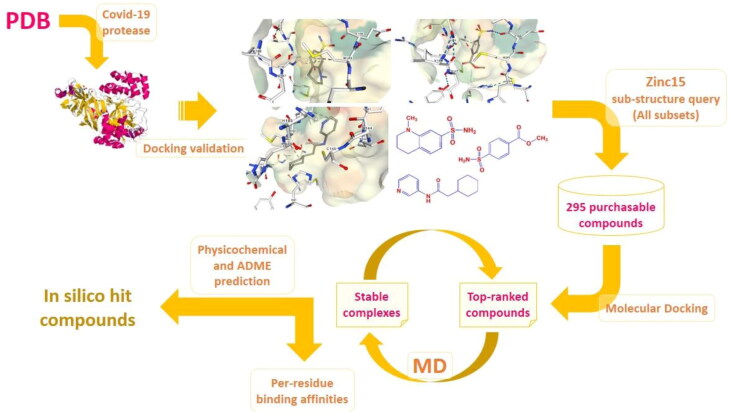
Hierarchical view of the multi-step simulations to identify hit compounds as potential SARS-CoV2 inhibitors; Covid-19 related 3 D Mpro structures were screened and subjected to AutoDock4.2 validation step to afford three holo structures with PDB IDs 5R80, 5R81 & 5R84 (Fearon et al., 2020). Corresponding co-crystallographic fragment ligands were subjected to substructure query in Zinc15 database with the criteria set on all subsets including purchasable compounds (Sterling & Irwin, 2015). 295 compounds were selected and entered into molecular docking study to attain three top-ranked analogues with highest binding affinities to related protease targets. Time dependent stability of top-ranked ligand-protease complexes were checked by MD simulations within 50 ns and the cycle might be repeated for next top-ranked candidates if non-desirable results were achieved. Subsequent amino acid decomposition analysis was performed on in silico hit compounds to acquire ligand-residue intermolecular binding energies by functional B3LYP in association with split valence basis set using polarization functions (Def2-TZVPP). Schematic 3 D representation of fragment-enzyme complexes were generated by NGL which is a WebGL based 3 D viewer incorporated into PDB (Rose et al., 2018).