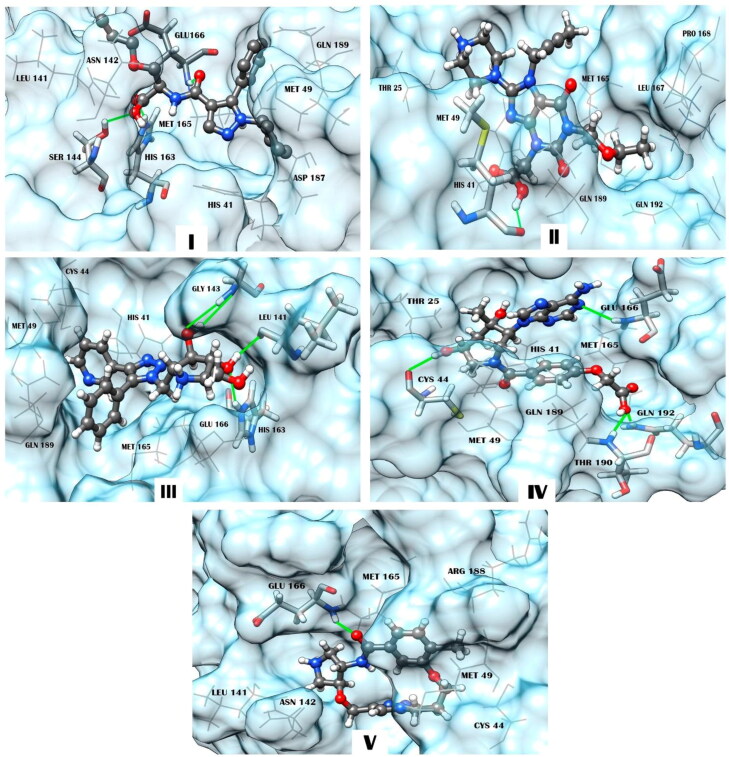
Figure 3.
Structural complexes of compounds I, II, III, IV, and V with SARS-CoV-2 MPro generated by molecular docking. The compounds are represented by a ball-stick-ball model. The enzyme residues forming interatomic contacts with the ligands are indicated (Table 4). Residues of MPro involved in hydrogen bonding are noted using a stick model. Hydrogen bonds are shown by solid lines. A wire model is used to designate residues forming van der Waals contacts, salt bridges, and π- or T-stacking.