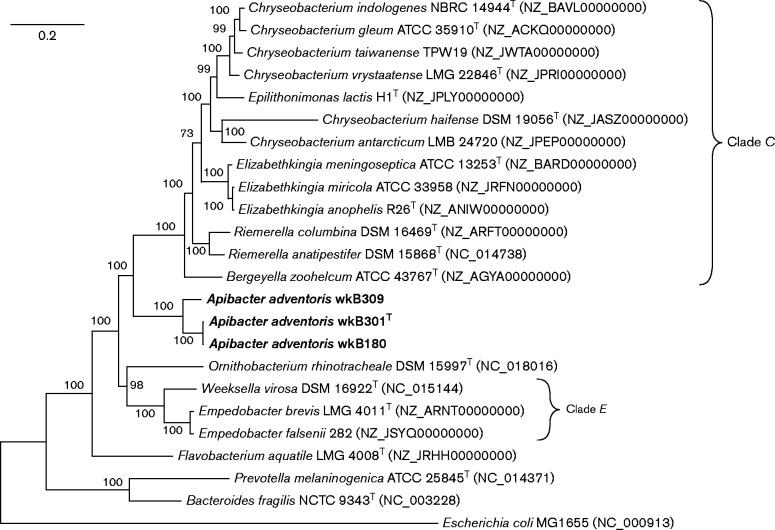
Fig. 2.
Phylogenetic tree of strains wkB301T, wkB309, wkB180 and related taxa, based on 20 conserved proteins (encoded by genes alaS, atpA, dnaA, dnaN, ftsZ, fusA, groEL, gyrB, lepA, metK, nusG, pfkA, pyrG, recA, rplA, rplB, rpoB, rpsB, sdhA and secA). Accession numbers for these genes are listed in Table S2. Proteins were concatenated and aligned with muscle and the tree was built with a maximum-likelihood algorithm using the Jones–Taylor–Thornton substitution model with gamma-distributed rates and invariant sites. Bootstrap percentages (1000 replicates) are shown at nodes. Bar, 0.2 substitutions per site.