Abstract
Background
Targeting immune checkpoint proteins has recently gained substantial attention due to the dramatic success of this strategy in clinical trials for some cancers. Inducible T-cell co-stimulator ligand (ICOSLG) is a member of the B7 family of immune regulatory ligands, expression of which in cancer is implicated in disease progression due to regulation of antitumor adaptive immunity. Although aberrant ICOSLG expression has been reported in glioma cells, the underlying mechanisms that promote glioblastoma (GBM) progression remain elusive.
Methods
Here, we investigated a causal role for ICOSLG in GBM progression by analyzing ICOSLG expression in both human glioma tissues and patient-derived GBM sphere cells (GSCs). We further examined its immune modulatory effects and the underlying molecular mechanisms.
Results
Bioinformatics analysis and GBM tissue microarray showed that upregulation of ICOSLG expression was associated with poor prognosis in patients with GBM. ICOSLG expression was upregulated preferentially in mesenchymal GSCs but not in proneural GSCs in a tumor necrosis factor-α/nuclear factor-kappaB–dependent manner. Furthermore, ICOSLG expression by mesenchymal GSCs promoted expansion of T cells that produced interleukin-10. Knockdown of the gene encoding ICOSLG markedly reduced GBM tumor growth in immune competent mice, with a concomitant downregulation of interleukin-10 levels in the tumor microenvironment.
Conclusions
Inhibition of the ICOSLG–inducible co-stimulator axis in GBM may provide a promising immunotherapeutic approach for suppressing a subset of GBM with an elevated mesenchymal signature.
Keywords: glioma stem cell, ICOSLG, immune escape, mesenchymal type, glioblastoma
Key Points.
1. ICOSLG expression was upregulated preferentially in mesenchymal GSCs.
2. Increased ICOSLG expression is associated with poor prognosis in patients with GBM.
Importance of the Study.
We have identified a novel molecular mechanism underlying the dysfunctional immune microenvironment of GBM. In contrast to proneural GBM, mesenchymal GBM expressed ICOSLG and specifically drove the generation of T cells producing interleukin-10. Our findings provide molecular evidence that mesenchymal GBM is a more immune-resistant phenotype.
Accumulating evidence suggests that the stem-like properties of glioblastoma (GBM) sphere cells (GSCs) contribute to therapeutic resistance in GBM. In general, GBM cells create an immunosuppressive microenvironment and employ various molecular and cellular alterations to escape immune surveillance. Wei et al have recently shown that GSCs are powerful mediators of immunosuppression of the adaptive immune system, specifically T-cell responses.1 A recent advance in GBM biology has divided GSCs into 2 subtypes, with the mesenchymal (MES) GSC population as the more malignant subtype.2 Proneural (PN) GSCs, on the other hand, induce significantly stronger transforming growth factor-β–mediated suppression of CD8+ T cells and natural killer (NK) cells.3 Interleukin (IL)-10 is a common prognostic marker in GBM. However, the underlying molecular mechanisms that regulate IL-10 production are poorly understood.4
Inducible T-cell co-stimulator ligand (ICOSLG), a member of the B7 family of ligands, plays an important role in regulatory T cell (Treg)–mediated immune responses. Our previous studies have revealed that expression of ICOSLG in plasmacytoid dendritic cells supports cancer progression by promoting the accumulation of immunosuppressive CD4+ T cells.5,6 Others have shown that ICOSLG expressed on gastric cancer cells and melanoma can induce the activation of IL-10–producing CD4+ T cells.7 GBM also expresses ICOSLG, but its role in tumor progression remains unclear.8
In this study, we have identified a novel molecular mechanism underlying the dysfunctional immune microenvironment of GBM. In contrast to PN GSCs, MES GSCs expressed ICOSLG and specifically resulted in the generation of IL-10–producing pro-tumorigenic T cells.
Materials and Methods
Detailed description of the materials and methods used in this study can be found in the Supplementary material.
Ethics
All work related to human tissues was approved by the institutional review board at Kansai Medical University (IRB #N1401), and subjects and/or their parents or guardians provided written informed consent. All mouse experiments were conducted under the approved protocols of the Institutional Animal Care and Use Committee at Kansai Medical University (IACUC #N16-119) and in accordance with guidelines from the National Institutes of Health.
GSC Cultures
High-grade glioma patient-derived neurospheres were established from surgical specimens obtained by Dr Nakano and his colleagues and molecularly characterized, as previously described.2,9–11 The murine RasR2 glioma-initiating cell line was generously provided by Dr Hideyuki Saya (Keio University, Japan).12
Flow Cytometry Analysis
The expression of B7 family proteins and class 1 and class 2 human leukocyte antigen (HLA) by GSC tumor cell lines was determined with flow cytometry (FACS Calibur system, BD Biosciences). The mean fluorescence intensity ratio (MFI-R) was calculated from all live cells according to the following formula: MFI of ICOSLG-stained sample histogram / MFI of the control histogram.
Immunofluorescence Staining
Mice brains were fixed in 4% paraformaldehyde, paraffin embedded, and cut into 10-µm sections. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 in phosphate buffered saline. Images were obtained using confocal microscopy (LSM510 META, Carl Zeiss).
Nuclear Factor-KappaB Promoter Luciferase Assay
The resulting vector was designated pNFκBΔRE-Luc. The luciferase activities were assessed using the Dual-Glo Luciferase Assay System (Promega). Luminescence was measured with a 2030 ARVO X Multilabel Reader (PerkinElmer).
Transfection of siRNA Oligonucleotides
Small interfering (si)RNA of NF-κBp65 or control siRNA (Cell Signaling) was transfected using Lipofectamine RNAiMAX (Invitrogen). After 24 h, the cells were treated with 10 ng/mL tumor necrosis factor-α (TNF-α).
Hypoxia Induction
GSCs were incubated in hypoxic conditions in an incubator with a gas mixture of N2/CO2/O2 (94:5:1) for 48 h.
Cytometry by Time-of-Flight Staining and Data Analysis
Cytometry by time-of-flight (CyTOF) was carried out as previously described.13 Cells were then stained and analyzed by CyTOF. The CyTOF data were exported in a conventional flow-cytometry file (.fcs) format and normalized using previously described software.14
Treatment of CD4+ Naïve T Cells with Medium Conditioned by GSCs
Tumor conditioned medium (CM) was prepared by culturing GSCs in Dulbecco’s modified Eagle’s medium/F12 for 24 h. CD4+ naïve T cells (purity >99%) were isolated from peripheral blood mononuclear cells using the CD4+ T-cell Isolation Kit II (Miltenyi Biotec), as previously described.15 Detailed protocols are available in the Supplementary methods.
Enzyme-Linked Immunosorbent Assay
The levels of ICOSLG and IL-10 were measured with enzyme-linked immunosorbent assay (ELISA) (MyBioSource).
ICOSLG Silencing
Lentiviral particles encoding 3 target-specific 19- to 25-nucleotide (plus hairpin) short hairpin (sh)RNAs (shICOSLG; sc-42768-V and sc-42769-V, Santa Cruz Biotechnology) were used to knock down ICOSLG expression.
Cell Proliferation Assay
Cell proliferation was determined with the Cell Counting Kit 8 (Dojindo). Normalized fold changes were calculated as the relative cell proliferation (% of control).
ICOSLG-Targeted Effects in Mouse Models of GBM
GSCs were stereotactically injected into the right striatum of athymic mice (Balb/c nu/nu) and/or immunocompetent mice (C57BL/6) (n = 6 per treatment group).
Quantitative Real-Time PCR
Quantitative (q)PCR was performed with the ABI 7300 system (Applied Biosystems; Thermo Fisher Scientific) by using the Thunderbird qPCR mix (Toyobo). The relative gene expression was calculated with the ΔΔCt method.
Patient Sample Analysis
Differential expression of ICOSLG was analyzed in GBM (n = 607) and normal unmatched brain samples (n = 11) from The Cancer Genome Atlas (TCGA) database. The correlation between ICOSLG expression and patient survival was assessed using cBioportal (www.cBioportal.org).
Immunohistochemistry
Surgical specimens from newly diagnosed GBM patients were obtained from the Kansai Medical University. All tissues were fixed in buffered formalin, embedded in paraffin, and stained with hematoxylin and eosin. Photomicrographs were obtained with a digital slide scanner (NanoZoomer-XR).
Data and Statistical Analysis
Statistical analyses were performed with EZR (Saitama Medical Center, Jichi Medical University).16 Data were analyzed with one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test.
Results
ICOSLG Is Preferentially Expressed in MES GSCs
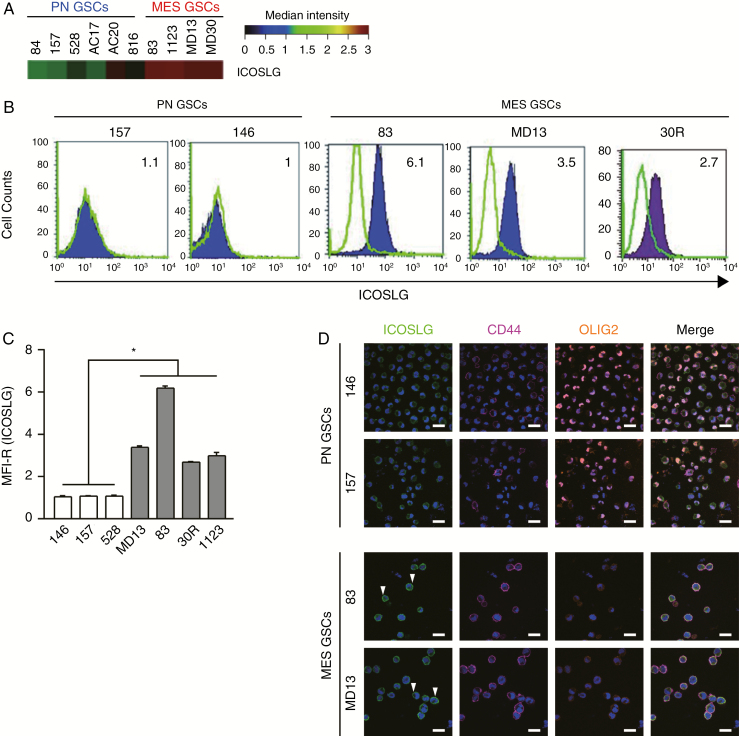
To investigate the tumorigenic potential of ICOSLG in GSCs, we first analyzed expression of ICOSLG mRNA in a panel of 10 patient-derived GSCs retrieved from our previously published microarray study.2,9–11 The heat map indicated that ICOSLG expression was significantly upregulated in MES type GSCs compared with PN type GSCs (Fig. 1A). To investigate the tumorigenic potential of ICOSLG-expressing tumor cells, we further analyzed expression of ICOSLG with flow cytometry in a panel of 7 patient-derived GBM sphere cultures, as described previously.9 Flow cytometry analysis and MFI-R (calculated considering all live cells according to the following formula: MFI of ICOSLG-stained sample histogram / MFI of the control histogram) quantitation indicated that ICOSLG protein levels were elevated in 4 MES GSC lines (MD13, 83, 30R, and 1123; Fig. 1B and C). In clear contrast, 3 PN GSC lines (146, 157, and 528) showed a marginal level of ICOSLG expression (Fig. 1B and C). Our results from fluorescence activated cell sorting (FACS) analysis were further confirmed with immunocytochemistry of MES (MD13 and 83) and PN (146 and 157) GSC lines. Higher expression of ICOSLG was associated with the MES marker CD44 compared with the PN marker OLIG2 (Fig. 1D). As positive controls for staining, we confirmed the presence of ICOSLG in human umbilical cord (Supplementary Fig. 1A, B) and mouse spleen (Supplementary Fig. 1C, D). In addition, we assessed expression of programmed cell death ligand 1 (PD-L1), HLA-ABC, and HLA-DR in these GSC lines with flow cytometry (Supplementary Table 1). All GSCs expressed PD-L1 and HLA-ABC, but not HLA-DR. Collectively, these data indicated that ICOSLG may be an additional valid therapeutic target in the MES subtype of GBM tumors. We next examined the possible target for therapy for the ICOSLG-ICOS axis in our preclinical models of GBM.
Fig. 1.
ICOSLG is highly expressed in MES GSCs. (A) ICOSLG mRNA expression was analyzed in MES and PN GSCs with transcriptomic microarray experiments. Total mRNA was extracted from GSCs and analyzed to create a gene expression profile. Results are expressed as the mRNA expression fluorescence intensity. (B) GSCs were stained with isotype control (green histograms) or ICOSLG-specific (purple histogram) antibodies and analyzed with flow cytometry. (C) The mean fluorescence intensity ratio (MFI-R) was calculated as the MFI of ICOSLG-positive sample histograms divided by the MFI of the control histogram. The MFI-Rs of the ICOSLG signal in the indicated GSCs are presented as the means ± standard error of 3 independent experiments (*P < 0.05, MES [n = 4] vs PN [n = 3], one-way ANOVA). (D) Immunofluorescent staining for ICOSLG expression in MES and PN glioma GSCs. The green fluorescence of ICOSLG on MES GSCs (arrowheads). The fluorescence of CD44 (purple) on the cell membrane. The fluorescence of OLIG2 (orange) in nuclei of PN GSCs. 4′,6′-Diamidino-2-phenylindole was used to stain nuclei (blue). Scale bars represent 20 μm.
Expression of ICOSLG Is Regulated by TNF-α/Nuclear Factor kappaB Signaling in GSCs
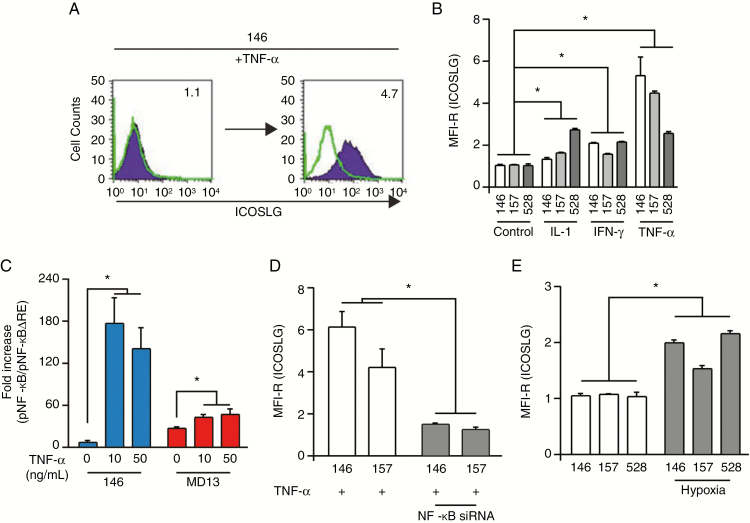
To understand the molecular mechanisms that regulate ICOSLG signaling in GBM, we investigated which proinflammatory cytokines (IL-1β, interferon [IFN]-γ, and TNF-α) activate ICOSLG expression in GSCs. Given that our prior studies identified TNF-α as a key regulator that promotes MES identity in GSCs, we tested the effects of IL-1β, IFN-γ, and TNF-α on PN GSCs.9,10,17 Treatment of the PN GSC lines (146, 157, and 528) with IL-1β, IFN-γ, or TNF-α at 10 ng/mL for 96 h induced expression of ICOSLG (Fig. 2A and B). Of these cytokines, TNF-α treatment showed the most prominent effect. When we compared nuclear factor kappaB (NF-κB) activity with luciferase assay between ICOSLGhigh MD13 MES GSCs and ICOSLGlow 146 PN GSCs, we found that MD13 had higher basal NF-κB activity (Fig. 2C). Interestingly, TNF-α treatment dramatically increased transcriptional activation of NF-κB in 146 PN GSCs, whereas such a stimulatory effect on NF-κB signaling was relatively less obvious in MD13 MES GSCs. Moreover, the maximum levels of NF-kB expression were different, suggesting qualitative differences between 146 PN and MD13 MES GSCs (Fig. 2C). We investigated whether NF-κB inhibition influences the expression of ICOSLG on PN GSCs with siRNA. Silencing NF-κBp65 significantly attenuated ICOSLG expression with TNF-α treatment (Fig. 2D). In addition, when PN GSCs were incubated in hypoxic conditions to induce MES trans-differentiation, ICOSLG expression was increased (Fig. 2E).
Fig. 2.
Expression of ICOSLG is regulated by NF-κB. (A)FACS analysis of ICOSLG expression in 146 PN GSCs after 96 h of treatment with 10 ng/mL TNF-α. GSCs were stained with isotype control (green histograms) or ICOSLG-specific (purple histogram) antibodies. The numbers in each panel indicate the MFI-R. (B) MFI-R of ICOSLG-expressing PN GSCs (146, 157, 528) analyzed after stimulation with proinflammatory cytokines (IL-1β, IFN-γ, or TNF-α) for 96 h (A). Data are expressed as the means ± standard error of three independent experiments (*P < 0.05, PN GSCs [n = 3] vs. PN GSCs + proinflammatory cytokines [n = 3], one-way ANOVA). (C) Luciferase-reported NF-κB activity was measured in MD13 (MES) and 146 (PN) GSCs. Data represent the means ± standard deviation (*P < 0.01, n = 3). (D)Silencing NF-κBp65 significantly attenuated ICOSLG expression with TNF-α treatment. Data are expressed as the means ± standard error of 3 independent experiments (*P < 0.05, PN GSCs [n = 3] vs. PN GSCs + siNF-κBp65 [n = 3], one-way ANOVA). (E) PN GSCs (146, 157, 528) were grown in 1% oxygen for 48 h, and the level of ICOSLG was determined with flow cytometry. Data are expressed as the means ± standard error of 3 independent experiments (*P < 0.05, PN GSCs [n = 3] vs PN GSCs + hypoxia [n = 3], one-way ANOVA).
Characterization of Tumor-Infiltrating T Cells in GBM
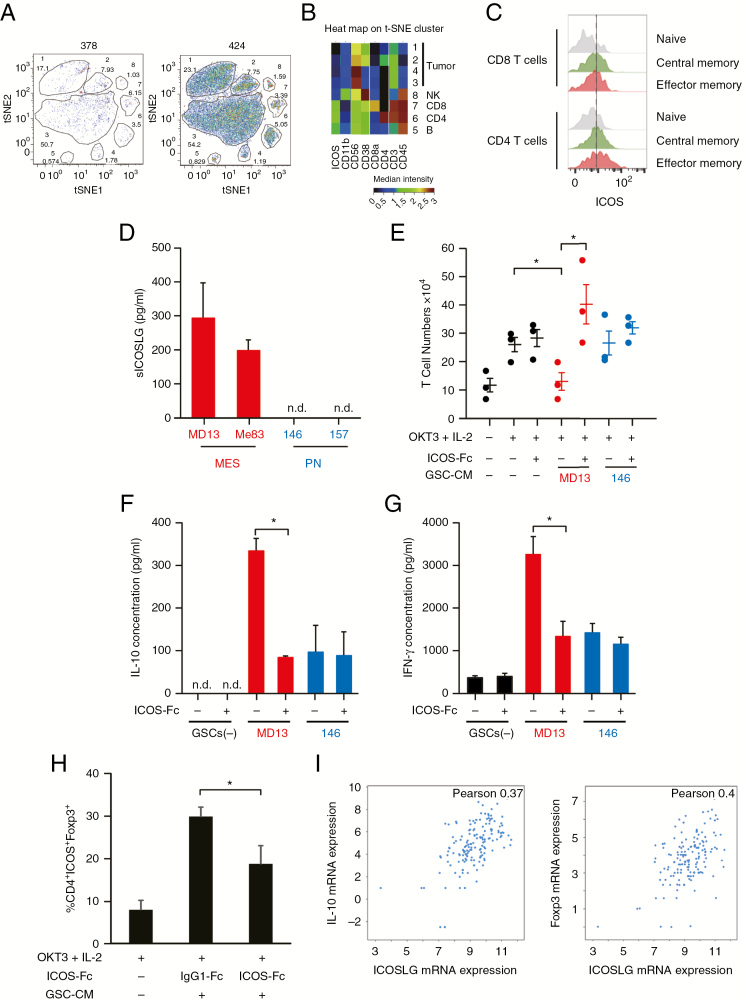
We first performed mass cytometry (CyTOF) to analyze the intratumoral immune profile in GBM tumor tissues. Peripheral blood mononuclear cells were used as a positive control. To identify the phenotype of tumor-infiltrating lymphocytes, a panel of 23 markers of identity, activation, and polarization was investigated, including CD56, CD39, OX40, CD38, PD1, 2B4, CD161, CD160, CD4, CD103, CD25, TIM-3, CD11b, ICOS, CD8a, CD8b, CD45, CD127, CD3, CCR7, CD45RO, CD28, and LAG3. CD45+ leukocytes were 10.8% of viable cells in the primary GBM tumor (424) and 11.8% in recurrent GBM (378) (Supplementary Fig. 2). Analysis by t-distributed stochastic neighbor embedding (t-SNE) of live cells showed 8 different clusters in GBM 378 and GBM 424 (Fig. 3A). Clusters 1–4 corresponded to tumor cells (CD45−). Clusters 5, 6, 7, and 8 corresponded to immune cells (CD45+). We identified CD8+ T cells (cluster 7), CD4+ T cells (cluster 6), and 2 other unidentified immune cell populations, including possibly NK (CD11b+/CD56+) cells (cluster 8) and B cells (cluster 5) (Fig. 3B). The results of CyTOF indicated that T cells were the main immune cell population that had infiltrated into GBM tumors (6.65% of total live cells). We then specifically analyzed expression of inducible co-stimulator (ICOS) on these T-cell populations. ICOS expression was observed both in effector memory CD4+ T cells and CD8+ T cells, but not in naïve T cells (Fig. 3C). Consistent with previous data, ICOS was predominantly expressed on CD4+ T cells relative to CD8+ T cells, especially before activation.18
Fig. 3.
Effects of ICOSLG on immune cells. (A) Representative image of t-SNE analysis of 378 GBM cells. Upper number: Cluster ID (1 to 8). Bottom number: Cluster frequency in live cells (Cluster 1, 17.1%; Cluster 2, 7.93%; Cluster 3, 1.03%; Cluster 4, 1.78%; Cluster 5, 0.574%; Cluster 6, 3.5%; Cluster 7, 6.15%). (B) Heat map of the t-SNE cluster (424 GBM cells). (C) ICOS expression on CD4+ and CD8+ T cells in a GBM patient (patient 424). Gray; naïve, green; central memory, red; effector memory. (D) Production of soluble ICOSLG by MES and PN GSCs. Data represent the mean ± standard error of triplicate experiments. (E) Naïve peripheral CD4+ T cells were cultured with or without tumor cell conditioned medium. The cells were incubated with IL-2 (50 IU/mL) and OKT3 (0.2 µg/mL) in the presence of neutralizing antibodies (ICOS-Fc; 2 µg/mL) against ICOSLG or isotype-matched control antibodies (immunoglobulin G1–Fc; minus) for 6 days. After 6 days of culture, T cells were collected and analyzed. T cells were quantified using trypan blue exclusion, and T-cell expansion was calculated. Each symbol represents an independent experiment performed in triplicate, and horizontal bars represent the mean. *P < 0.05, Student’s t-test. (F) The ability of primed T cells to secrete IL-10 was assayed with ELISA. The cell number was adjusted before polyclonal restimulation with anti-CD3 (5 µg/mL) and anti-CD28 (1 µg/mL) monoclonal antibodies (mAbs) and supernatants were collected 24 h after. Data represent the mean ± standard error of independent experiments performed in triplicate (*P < 0.05, Student’s t-test). n.d. indicates that the measured value was below the detection limit of the assay (<20 pg/mL). (G) The ability of primed T cells to secrete IFN-γ was assayed with ELISA. The cell number was adjusted before polyclonal restimulation with anti-CD3 (5 µg/mL) and anti-CD28 (1 µg/mL) mAbs and supernatants were collected 24h after. Data represent the mean ± standard error of independent experiments performed in triplicate (*P < 0.05, Student’s t-test). n.d. indicates that the measured value was below the detection limit of the assay (<20 pg/mL). (H) CD4+ T cells cultured with MES GSC conditioned medium in the presence of ICOS-Fc or immunoglubulin G1–Fc were analyzed for the expression of ICOS and Foxp3 with flow cytometry. The percentages of ICOS+Foxp3+ cells in the CD4+ population are shown. Data represent the mean ± standard error of independent experiments performed in triplicate (*P < 0.05, Student’s t-test). (I) RNAseq data from n = 206 GBM patient samples (TCGA Research Network) were used to compare IL-10 and Foxp3 expression with expression of ICOSLG. Visualization of the data was performed using cBioportal.
ICOSLG Expression in MES GSCs Induced Proliferation of IL-10–Producing T Cells
Next, we evaluated the functional consequences of ICOSLG stimulation on human GSCs. Our previous study reported that ICOSLG induced IL-10 production in T cells, which could promote Treg-mediated peripheral tolerance.19 The soluble form of ICOSLG (sICOSLG) is functional,20 and hence the levels of sICOSLG in MES and PN GSCs were measured with ELISA. Our results indicated that only CM derived from MES GSCs (MD13 and 83) showed detectable levels of sICOSLG (Fig. 3D). Thus, to assess the role of ICOSLG expression by MES GSCs in the priming of T-cell responses, we analyzed the effect of adding recombinant ICOSLG protein on the generation of IL-10–producing T cells by MES GSCs. We then cultured allogeneic peripheral blood-derived naïve CD4+ T cells together with tumor CM. To determine a role for ICOSLG in T cells, we blocked ICOS co-stimulation with an ICOS-Fc decoy protein against sICOSLG. MES GSC-CM inhibited expansion of naïve T cells by IL-2/OKT3(CD3) stimulation (Fig. 3E). However, the ICOS-Fc decoy protein restored the expansion of naïve T cells, suggesting a role for ICOSLG in blockade of primary T-cell activation (Fig. 3E). To determine whether ICOSLG on MES GSCs is involved in the polarization of naïve T cells, we tested the ability of primed T cells to secrete cytokines in response to polyclonal restimulation. T cells primed with CM from MD13 MES GSCs produced a high level of IL-10 (296–403 pg/mL) (Fig. 3F) and IFN-γ (2520–4220 pg/mL) (Fig. 3G), whereas IL-4 secretion was limited (data not shown). Incubation with CM from MES GSCs upregulated the number of ICOS- and Foxp3-double-positive cells, whereas co-treatment with ICOS-Fc significantly decreased the indicated subpopulation of cells (Fig. 3H). These results were consistent with TCGA Research Network data, which showed that expression of both IL-10 and Foxp3 mRNA in GBM tumors was positively correlated with expression of ICOSLG (Fig. 3I). Collectively, these data suggest that sICOSLG released from MES GBM cells induces CD4+ T cell expansion and IL-10 production by these cells.
Effects of ICOSLG Silencing in Preclinical GBM Mouse Models
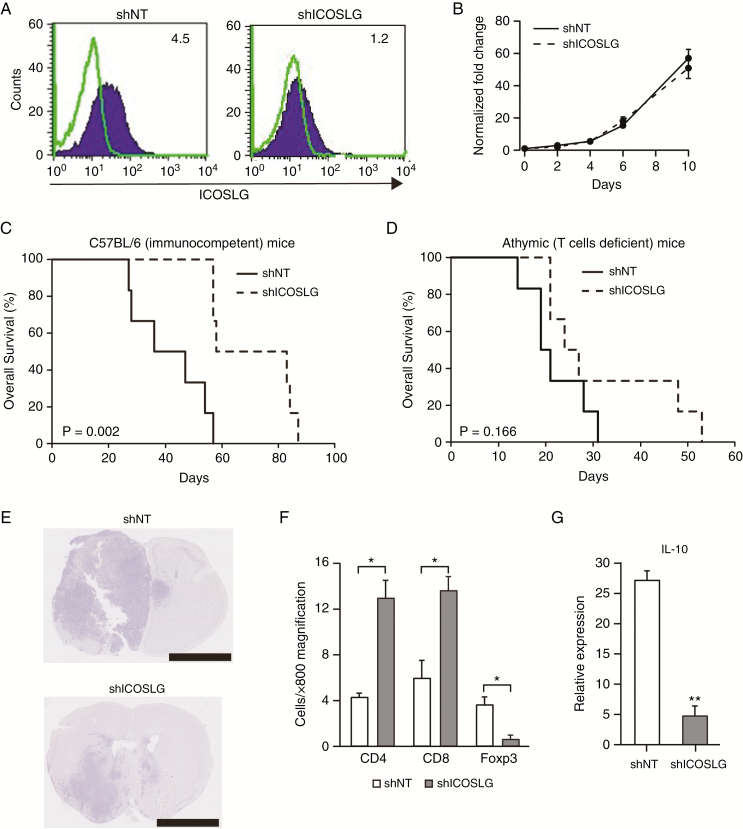
We then used an immunocompetent mouse model of GBM to further understand the role of immune cells in the action of ICOSLG on GBM aggressiveness. To this end, we used mouse glioma cancer-initiating cells (RasR2).12 We first confirmed ICOSLG expression in RasR2 cells with flow cytometry (Fig. 4A). In contrast, expression of ICOSLG was not detected in GL261, a mouse glioma cell line. Stable induction of ICOSLG knockdown was established in RasR2 cells using an shRNA construct targeting ICOSLG. Flow cytometry further confirmed downregulation of ICOSLG expression in RasR2-shICOSLG cells (Fig. 4A). Silencing of ICOSLG did not show a significant effect on the growth of RasR2 cells in vitro (Fig. 4B). In contrast to this in vitro data, in the immunocompetent mouse model (C57BL/6), silencing of ICOSLG in RasR2 cells significantly prolonged the median survival of tumor-bearing mice (P = 0.002 for shICOSLG; log-rank P-value, Fig. 4C). In contrast, the survival benefit of ICOSLG silencing in RasR2 cells was largely eliminated in T-cell deficient athymic mice (n = 6 per group, P = 0.166 for shICOSLG; log-rank P-value; Fig. 4D). Although shICOSLG had a nonsignificant effect on the in vitro growth of RasR2 cells, it decreased in vivo tumor growth compared with the scrambled control, shNT (Fig. 4E). We further characterized the in vivo effects of ICOSLG silencing on tumor-infiltrating T-cell populations by analyzing GBM tumor sections. To investigate the cellular basis for the immunosuppressive effect of ICOSLG, we analyzed the presence of Foxp3+, CD4+, and CD8+ T cells in the brain tumor microenvironment at 30 days post-RasR2 transplantation. Knockdown of ICOSLG caused a significant decrease in the number of Foxp3+ cells compared with the NT control (Fig. 4F, Supplementary Fig. 4). Notably, the numbers of CD4+ and CD8+ T cells in RasR2-shICOSLG tumor sections were significantly higher than those from RasR2-shNT tumors (Fig. 4F, Supplementary Fig. 4). To further investigate whether ICOSLG silencing modulates the immune response in the brain tumor microenvironment, the levels of IL-10 mRNA expression were evaluated in RasR2-shICOSLG tumors with real-time PCR. Silencing of ICOSLG downregulated IL-10 expression compared with control mice (P < 0.01; Fig. 4G). Taken together, the results of preclinical GBM models indicated a pro-tumorigenic role for ICOSLG in GBM, and implicated interactions between ICOSLG+ GSCs and ICOS+ Tregs as a potential mechanism of GBM progression.
Fig. 4.
Effects of ICOSLG knockdown in vivo in a syngeneic brain tumor model. (A) Representative flow cytometry results showing ICOSLG expression in mouse RasR2 GSCs with stable induction of shICOSLG or shNT expression. The numbers in each panel indicate the MFI-R. (B) Effects of ICOSLG knockdown with shICOSLG on cell proliferation in RasR2 glioma spheres using Cell Counting Kit 8. (C) Kaplan–Meier survival analysis evaluating the correlation between shICOSLG and shNT in immunocompetent mice (C57BL/6) (n = 6 per group, P = 0.002, log-rank test). (D) Kaplan–Meier survival analysis evaluating the correlation between shICOSLG and shNT in athymic mice (Balb/c nu/nu) (n = 6 per group, shICOSLG vs shNT in athymic mice; Balb/c nu/nu mice, P = 0.166, log-rank test). (E) Representative images of hematoxylin and eosin staining of mouse brains harvested on day 30 after transplantation of RasR2 glioma spheres expressing shNT or shICOSLG. Scale bars, 2.5 mm. (F) Numbers of Foxp3+, CD4+, and CD8+ T cells were counted in RasR2 tumor sections from the identical C57BL/6 mouse model (6 fields at 800× magnification, means ± standard error, *P < 0.05). (G) RNA was extracted from RasR2 tumors in brains of C57BL/6 mice and used for real-time PCR. The level of IL-10 expression was normalized to the level of Hprt (mean ± standard error, **P < 0.01, n = 3–5).
ICOSLG Expression in GBM Patients Was Inversely Correlated with Patient Survival
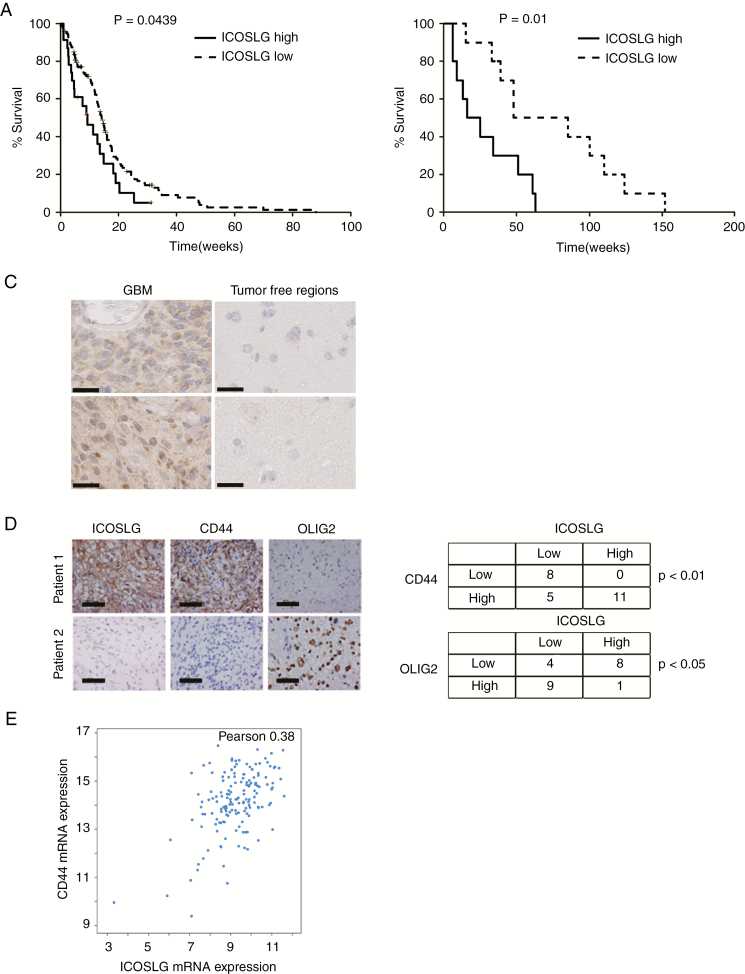
To determine the clinical significance of ICOSLG, we first investigated whether ICOSLG expression is correlated with the prognosis of GBM patients. Analysis of RNA-sequencing (RNAseq) data of GBM samples from TCGA Research Network using cBioPortal and the published dataset classifications21,22 revealed that higher expression of ICOSLG (Z-score >1.0) was significantly correlated with worse prognosis (log-rank P = 0.0439, ICOSLGhigh vs ICOSLGlow; Fig. 5A). This patient data analysis was further confirmed by immunohistochemical (IHC) assessment of 20 tumor sections from newly diagnosed GBM patients undergoing neurosurgical resection in the Department of Neurosurgery at Kansai Medical University. Similar to the results from TCGA data analysis, immunoreactivity to ICOSLG within these GBM tumors was negatively correlated with patient survival (log-rank P = 0.01, negative vs positive; Fig. 5B). Further, ICOSLG-expressing cells were prevalent throughout the GBM tumor sections, whereas ICOSLG expression in nontumor tissues was below the detectable level (Fig. 5C).
Fig. 5.
ICOSLG was highly expressed in CD44-high GBMs and predicted poor prognosis. (A) Analysis of TCGA database indicated an inverse correlation between ICOSLG mRNA expression and survival of GBM patients (P = 0.0439, log-rank test, ICOSLG upregulated [solid line] Z-score >1.0, n = 29 vs ICOSLG downregulated [broken line] Z-score <1.0, n = 42). (B) Kaplan–Meier analyses were used to evaluate the correlation between ICOSLG protein expression and survival of 20 newly diagnosed GBM patients (ICOSLG high vs low, P = 0.01, log-rank test). (C) Representative IHC images show the expression of ICOSLG in nontumor brain samples and GBM tissues (n = 2 per group). Scale bars, 50 μm. (D) Representative IHC images of ICOSLG, CD44, and OLIG2 expression in human GBM samples are shown. Scale bars, 50 μm. The tables show the number of patient samples that were classified according to the expression level of ICOSLG, CD44, and OLIG2. ICOSLG was positively correlated with CD44 but inversely correlated with OLIG2. (E) The expression level of mRNA with RNAseq from GBM patient samples (n = 206; TCGA Research Network). Pearson correlation of the expression between ICOSLG and CD44 is shown. Data were visualized using cBioportal.
ICOSLG Was Preferentially Expressed in MES GBMs (CD44high GBM)
Given that malignant progression in cancers is often associated with a gain of the MES phenotype, we next determined if ICOSLG expression was correlated with expression of CD44, a representative MES marker. IHC staining was carried out for ICOSLG, OLIG2 (a surrogate PN GBM marker), and CD44 (a surrogate MES GBM marker) in 24 newly diagnosed GBM specimens. As shown in Fig. 5D, ICOSLG expression was highly correlated with CD44 expression (P < 0.01, ICOSLGhigh vs ICOSLGlow), whereas expression of ICOSLG and OLIG2 were negatively correlated (P < 0.05). Consistent with our results from IHC, the RNAseq data generated by TCGA Research Network also showed a positive correlation between CD44 and ICOSLG mRNA expression in GBM patient specimens (P < 0.05, Pearson: 0.38, n = 206; Fig. 5E).
Discussion
In this study, we report that glioblastoma cells with high levels of ICOSLG expression are more tumorigenic in vivo than cells with low levels of ICOSLG. This is further supported by our analyses of clinical GBM patient samples. Another novel set of findings in this study is that GBM with the MES phenotype predominantly expresses ICOSLG, which is regulated by the NF-κB pathway. Although basal levels of ICOSLG expression were low in GSCs with the PN phenotype, stimulation with TNF-α substantially induced ICOSLG expression. We also demonstrated that knockdown of ICOSLG inhibits expansion of IL-10–producing T-cell subpopulations.
MES GBMs exhibit a high degree of macrophage/microglial infiltration, and the macrophages and microglia cells may provide extrinsic signals to promote the proneural-to-mesenchymal transition (PMT) through NF-κB activation.17 ICOSLG expression is highly elevated in the CD44high MES subtype of GBM and is informative for the prognosis of patients. Moreover, we report that ICOSLG was overexpressed in MES but not in PN GSCs. MES GSCs are the more aggressive and radiotherapy-resistant subtype.9–11 Our current study also reveals ICOSLG as a potential link between tumor immunosuppression and PMT in GSCs. Macrophages and microglia may provide extrinsic signals including TNF-α to cause ICOSLG activation, thereby promoting PMT of tumors and/or GSCs via NF-κB activation. Consistently, microglia and macrophage accumulation promotes tumor development after transplantation of RasR2 cells.12 TNF-α regulates the expression of ICOSLG on CD34+ progenitor cells during differentiation into antigen-presenting cells.23 This implies that tumor-infiltrating macrophages and microglia enhance ICOSLG expression on GSCs through TNF-α/NF-κB activation. TNF-α induces elevation of genes that are involved in wound healing.17 In addition to the immune response, ICOSLG signaling plays pivotal roles in skin wound healing via IL-6.24 IL-6 signaling is also important for initiation and progression of GBM. Moreover, Dianzani et al reported that ICOS-Fc inhibits epithelial-mesenchymal transition and migration in vitro as well as metastasis in vivo.25 In the present study, silencing of ICOSLG in RasR2 cells tended to prolong the median survival of T cell–deficient athymic mice (Fig. 4D). This preliminary observation implies that ICOSLG is involved in non-immunological functions, as well as IL-10–producing T-cell subpopulations.
Because of the central role of GSCs in glioblastoma initiation, progression, recurrence, resistance to chemotherapy and radiation, and immune suppression, GSCs have recently garnered interest as an immune therapeutic target. GSCs also exert a higher inhibitory effect on T-cell proliferation compared with the non-GSC population.26 We show here that MES GSCs suppressed proliferation of activated CD4+ T cells. We also observed that MES GSC-CM with ICOSLG blockade by ICOS-Fc increased the number of T cells compared with control (Fig. 3E) and that MD13 MES GSC-CM produced IL-10 (Fig. 3F) and induced CD4+ICOS+Foxp3+ T cells. Thus, we consider that even MES GSC-CM has the potential to induce proliferation of T cells with ICOS-Fc and that ICOSLG stimulation of MES GSCs during the culture process can induce IL-10 production, resulting in the suppression of proliferation of the entire T-cell population at the end of the culture period. We interpret our results to indicate that in our experimental setting, IL-10–producing T cells were generated from naïve T cells via proliferation in the early phase of the culture period. However, eventually, the IL-10 produced by the generated Tregs suppressed proliferation of the entire T-cell population in our experiments. A similar result was reported by Wang et al.27 Collectively, these results may lead to development of effective molecular immunotherapy. Tregs, which are characterized by the expression of Foxp3, can suppress antitumor immune responses.6,28 Previous studies have revealed that the number of Tregs increases within tumors and in the circulation of cancer patients.29 The percentage of tumor-infiltrating Tregs is shown to be correlated with the World Health Organization grade of brain tumors.30 Tregs, which expresses IL-17, were found in high-grade glioma tissue.31 ICOS is involved in the co-stimulation of T cells and is expressed only on activated T cells. Upon activation, Tregs also express co-stimulatory receptors, including ICOS and CD28, which further boost their activation, proliferation, and survival. ICOS+ Tregs suppress the function of dendritic cells in an IL-10-dependent manner.32 In GBM, circulating ICOS+ Tregs are present in higher numbers than in low-grade glioma.33 However, the source of ICOS co-stimulation of Tregs is largely unknown. The results of our study strongly indicated that MES GSCs express ICOSLG, which promotes expansion of IL-10–producing Tregs. The IL-10+IFN-γ+ T cell profile induced by ICOSLG is reminiscent of Treg type 1 cells.5,34 Inhibition of ICOS co-stimulation enhances the expansion of T cells, suggesting that IL-10–producing T cells are poorly proliferative, which is a cardinal feature of Treg cells. IL-10 is one of the most important prognostic markers in GBM progression. IL-10–producing T cells induced by ICOSLG have characteristics of Tregs, including the ability to suppress bystander T-cell activation through the secretion of IL-10.5 However, the underlying molecular and cellular mechanisms regulating IL-10 expression are poorly understood.4 Previous study demonstrated that ICOS signaling allowed the expression of Foxp3 and the production of IL-10, while blockade of ICOSLG during T-cell activation reduced Foxp3 expression.35 Here, we show that expression of ICOSLG on MES GSCs was associated with IL-10 production in the tumor microenvironment. On the other hand, PN GSCs caused a significantly stronger transforming growth factor-β-mediated suppression of CD8+ T cells and NK cells.3
Although ICOSLG is a potentially promising candidate target for suppressing MES GSCs, we should consider that excessive and prolonged ICOSLG inhibition may be detrimental because of its suppression of antitumor immunity. ICOS could be expressed on activated T cells, and ICOS expression is upregulated on IFN-γ–secreting T cells. Schreiner et al reported that ICOS signaling is necessary for the production of IFN-γ by T cells stimulated with ICOSLG-expressing glioblastoma cell lines.8 However, ICOS-deficient patients have decreased IL-10–producing T cells but normal numbers of T-helper type 2 cells and IFN-γ–producing T cells.36 These reports suggest that treatment that neutralizes ICOS with an anti-ICOS monoclonal antibody needs to be restricted to a short period, otherwise effector cells that express ICOS will be restored. Several studies using ICOSLG-transfected solid tumor cell lines found that ICOSLG induces CD8+ cytotoxic lymphocyte-mediated tumor regression.37,38 Conversely, Tamura et al reported that the expression of ICOSLG in leukemic cells is associated with a shorter overall survival in acute myeloid leukemia patients.39 A recent study revealed that administration of soluble ICOSLG-Fc delays tumor growth in mouse hematological neoplasms in syngeneic hosts.40 CD8+ T cells play a very important role in eliminating tumor cells from the body. The frequency of CD8+ T cells in tumor tissue can be a prognostic sign of tumor progression. Knockdown of ICOSLG on GSCs promoted the frequency of CD8+ T cells in tumor tissue and significantly prolonged the survival of GBM-bearing mice. Thus, removal of such negative factors in CD8+ T cells may maintain more CD8+ T cells in tumor tissues and may facilitate the elimination of tumors from the body. According to this hypothesis, a phase I clinical trial study (MEDI-570) has been initiated in which anti-ICOS monoclonal antibody is being used to treat patients with peripheral T-cell lymphoma follicular variant or angioimmunoblastic T-cell lymphoma.
In conclusion, we identified a novel molecular mechanism underlying the dysfunctional immune microenvironment of GBM. In contrast to PN GBM, MES GBM expresses ICOSLG and specifically drives the generation of IL-10–producing T cells. Our findings provide molecular evidence that MES GBM is a more immune-resistant phenotype, and further suggest that targeting the ICOS/ICOSLG pathway may restore antitumor immunity via reduced IL-10 production.
Supplementary Material
Acknowledgments
We thank Ms. Hitomi Tatsumi for invaluable secretarial assistance.
Conflict of interest statement. There are no conflicts of interest to declare.
Authorship statement. R. Iwata performed analysis on all samples, interpreted the data, wrote the manuscript, and acted as corresponding author. All authors contributed to the supervised development of the work and helped with data interpretation and manuscript evaluation. Conception and design: R. Iwata. Development of methodology: R. Iwata. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): R. Iwata, M. Nonaka, M. Maruyama, S. Oe, K. Ofune, Kyung-Don Kang, T. Ito, K. Yoshimura, T. Hashiba. Analysis and interpretation of data (eg, statistical analysis, biostatistics, computational analysis): R. Iwata. Writing, review, and/or revision of the manuscript: R. Iwata, J.H.L., M. Hayashi, I. Nakano, L. Norian. Administrative, technical, or material support (ie, reporting or organizing data, constructing databases): U. Dianzani, I. Nakano, Y. Nakano. Study supervision: I. Nakano, A. Asai.
Funding
This work was supported by the Japan Society for the Promotion of Science KAKENHI (15K19983, 15K10346, 18K08983, and 19K18408), research grant D1 from Kansai Medical University, and the Associazione Italiana Ricerca sul Cancro (IG 20714, AIRC, Milano).
References
- 1. Wei J, Barr J, Kong LY, et al. . Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol Cancer Ther. 2010;9(1):67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cheng P, Wang J, Waghmare I, et al. . FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res. 2016;76(24):7219–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beier CP, Kumar P, Meyer K, et al. . The cancer stem cell subtype determines immune infiltration of glioblastoma. Stem Cells Dev. 2012;21(15):2753–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng W, Ren X, Zhang C, et al. . Bioinformatic profiling identifies an immune-related risk signature for glioblastoma. Neurology. 2016;86(24):2226–2234. [DOI] [PubMed] [Google Scholar]
- 5. Ito T, Yang M, Wang YH, et al. . Plasmacytoid dendritic cells prime IL-10-producing T regulatory cells by inducible costimulator ligand. J Exp Med. 2007;204(1):105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Conrad C, Gregorio J, Wang YH, et al. . Plasmacytoid dendritic cells promote immunosuppression in ovarian cancer via ICOS costimulation of Foxp3(+) T-regulatory cells. Cancer Res. 2012;72(20):5240–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen XL, Cao XD, Kang AJ, Wang KM, Su BS, Wang YL. In situ expression and significance of B7 costimulatory molecules within tissues of human gastric carcinoma. World J Gastroenterol. 2003;9(6):1370–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schreiner B, Wischhusen J, Mitsdoerffer M, et al. . Expression of the B7-related molecule ICOSL by human glioma cells in vitro and in vivo. Glia. 2003;44(3):296–301. [DOI] [PubMed] [Google Scholar]
- 9. Mao P, Joshi K, Li J, et al. . Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110(21):8644–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim SH, Ezhilarasan R, Phillips E, et al. . Serine/threonine kinase MLK4 Determines mesenchymal identity in glioma stem cells in an NF-κB-dependent manner. Cancer Cell. 2016;29(2):201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sadahiro H, Kang KD, Gibson JT, et al. . Activation of the receptor tyrosine kinase AXL regulates the immune microenvironment in glioblastoma. Cancer Res. 2018;78(11):3002–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sampetrean O, Saga I, Nakanishi M, et al. . Invasion precedes tumor mass formation in a malignant brain tumor model of genetically modified neural stem cells. Neoplasia. 2011;13(9):784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simoni Y, Fehlings M, Kløverpris HN, et al. . Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity. 2017;46(1):148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finck R, Simonds EF, Jager A, et al. . Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83(5):483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ogata M, Ito T, Shimamoto K, et al. . Plasmacytoid dendritic cells have a cytokine-producing capacity to enhance ICOS ligand-mediated IL-10 production during T-cell priming. Int Immunol. 2013;25(3):171–182. [DOI] [PubMed] [Google Scholar]
- 16. Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48(3):452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bhat KPL, Balasubramaniyan V, Vaillant B, et al. . Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McAdam AJ, Chang TT, Lumelsky AE, et al. . Mouse inducible costimulatory molecule (ICOS) expression is enhanced by CD28 costimulation and regulates differentiation of CD4+ T cells. J Immunol. 2000;165(9):5035–5040. [DOI] [PubMed] [Google Scholar]
- 19. Ito T, Hanabuchi S, Wang YH, et al. . Two functional subsets of FOXP3+ regulatory T cells in human thymus and periphery. Immunity. 2008;28(6):870–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nazzal D, Gradolatto A, Truffault F, Bismuth J, Berrih-Aknin S. Human thymus medullary epithelial cells promote regulatory T-cell generation by stimulating interleukin-2 production via ICOS ligand. Cell Death Dis. 2014;5:e1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cerami E, Gao J, Dogrusoz U, et al. . The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Richter G, Hayden-Ledbetter M, Irgang M, et al. . Tumor necrosis factor-alpha regulates the expression of inducible costimulator receptor ligand on CD34(+) progenitor cells during differentiation into antigen presenting cells. J Biol Chem. 2001;276(49):45686–45693. [DOI] [PubMed] [Google Scholar]
- 24. Maeda S, Fujimoto M, Matsushita T, Hamaguchi Y, Takehara K, Hasegawa M. Inducible costimulator (ICOS) and ICOS ligand signaling has pivotal roles in skin wound healing via cytokine production. Am J Pathol. 2011;179(5):2360–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dianzani C, Minelli R, Gigliotti CL, et al. . B7h triggering inhibits the migration of tumor cell lines. J Immunol. 2014;192(10):4921–4931. [DOI] [PubMed] [Google Scholar]
- 26. Di Tomaso T, Mazzoleni S, Wang E, et al. . Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin Cancer Res. 2010;16(3):800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang W, Hodkinson P, McLaren F, et al. . Small cell lung cancer tumour cells induce regulatory T lymphocytes, and patient survival correlates negatively with FOXP3+ cells in tumour infiltrate. Int J Cancer. 2012;131(6):E928–E937. [DOI] [PubMed] [Google Scholar]
- 28. Strauss L, Bergmann C, Szczepanski M, Gooding W, Johnson JT, Whiteside TL. A unique subset of CD4+CD25highFoxp3+ T cells secreting interleukin-10 and transforming growth factor-beta1 mediates suppression in the tumor microenvironment. Clin Cancer Res. 2007;13(15 Pt 1):4345–4354. [DOI] [PubMed] [Google Scholar]
- 29. Lowther DE, Hafler DA. Regulatory T cells in the central nervous system. Immunol Rev. 2012;248(1):156–169. [DOI] [PubMed] [Google Scholar]
- 30. Sayour EJ, McLendon P, McLendon R, et al. . Increased proportion of FoxP3+ regulatory T cells in tumor infiltrating lymphocytes is associated with tumor recurrence and reduced survival in patients with glioblastoma. Cancer Immunol Immunother. 2015;64(4):419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang H, Yi L, Wang X, Zhou C, Xu L. Interleukin-17 facilitates the immune suppressor capacity of high-grade glioma-derived CD4 (+) CD25 (+) Foxp3 (+) T cells via releasing transforming growth factor beta. Scand J Immunol. 2014;80(2):144–150. [DOI] [PubMed] [Google Scholar]
- 32. Jacobs JF, Idema AJ, Bol KF, et al. . Prognostic significance and mechanism of Treg infiltration in human brain tumors. J Neuroimmunol. 2010;225(1-2):195–199. [DOI] [PubMed] [Google Scholar]
- 33. Gousias K, von Ruecker A, Voulgari P, Simon M. Phenotypical analysis, relation to malignancy and prognostic relevance of ICOS+T regulatory and dendritic cells in patients with gliomas. J Neuroimmunol. 2013;264(1-2):84–90. [DOI] [PubMed] [Google Scholar]
- 34. Groux H, O’Garra A, Bigler M, et al. . A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389(6652):737–742. [DOI] [PubMed] [Google Scholar]
- 35. Martin-Orozco N, Li Y, Wang Y, et al. . Melanoma cells express ICOS ligand to promote the activation and expansion of T-regulatory cells. Cancer Res. 2010;70(23):9581–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Warnatz K, Bossaller L, Salzer U, et al. . Human ICOS deficiency abrogates the germinal center reaction and provides a monogenic model for common variable immunodeficiency. Blood. 2006;107(8):3045–3052. [DOI] [PubMed] [Google Scholar]
- 37. Liu X, Bai XF, Wen J, et al. . B7H costimulates clonal expansion of, and cognate destruction of tumor cells by, CD8(+) T lymphocytes in vivo. J Exp Med. 2001;194(9):1339–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wallin JJ, Liang L, Bakardjiev A, Sha WC. Enhancement of CD8+ T cell responses by ICOS/B7h costimulation. J Immunol. 2001;167(1):132–139. [DOI] [PubMed] [Google Scholar]
- 39. Tamura H, Dan K, Tamada K, et al. . Expression of functional B7-H2 and B7.2 costimulatory molecules and their prognostic implications in de novo acute myeloid leukemia. Clin Cancer Res. 2005;11(16):5708–5717. [DOI] [PubMed] [Google Scholar]
- 40. Ara G, Baher A, Storm N, et al. . Potent activity of soluble B7RP-1-Fc in therapy of murine tumors in syngeneic hosts. Int J Cancer. 2003;103(4):501–507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.