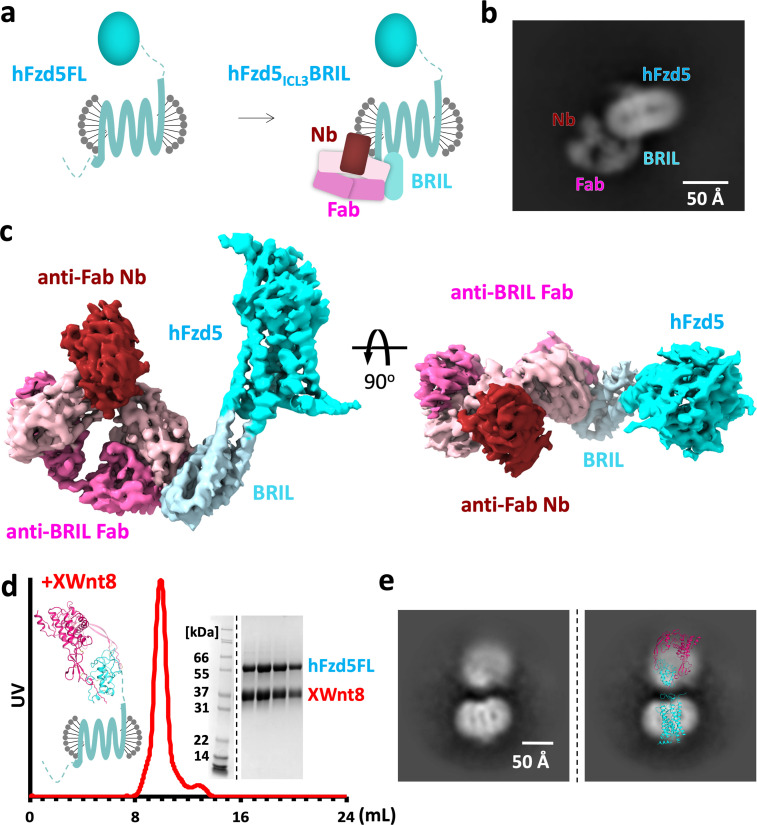
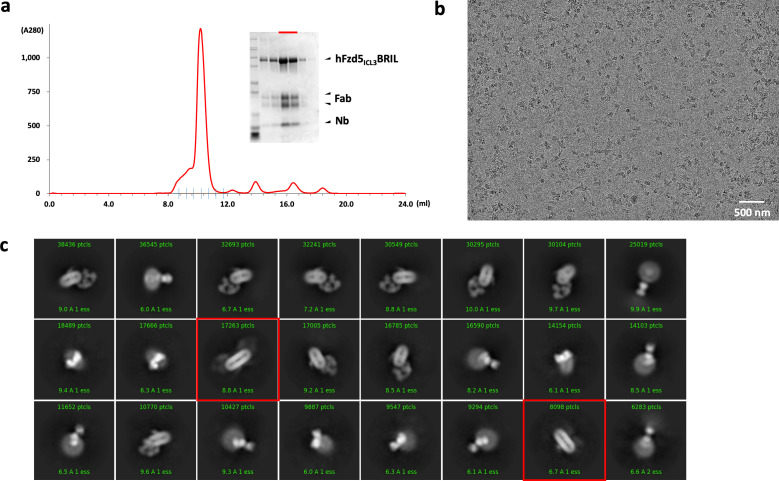
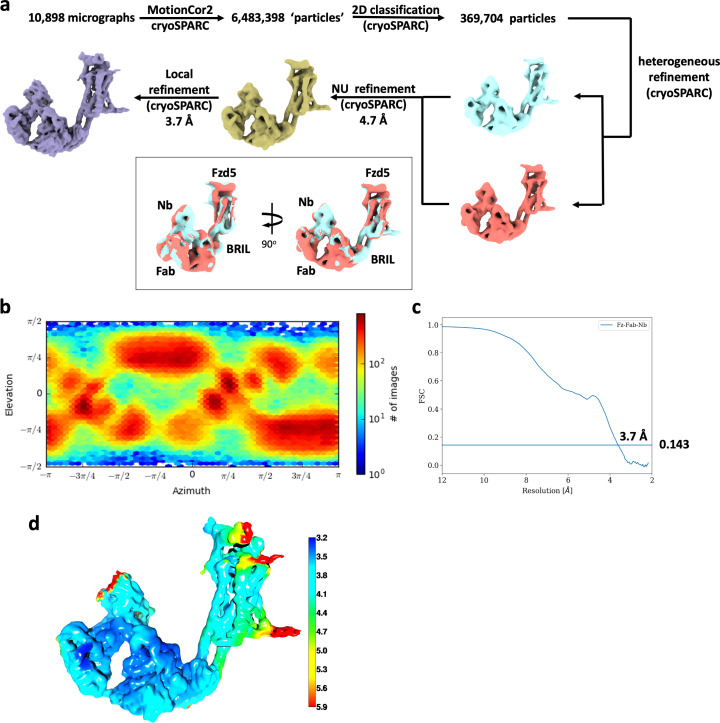
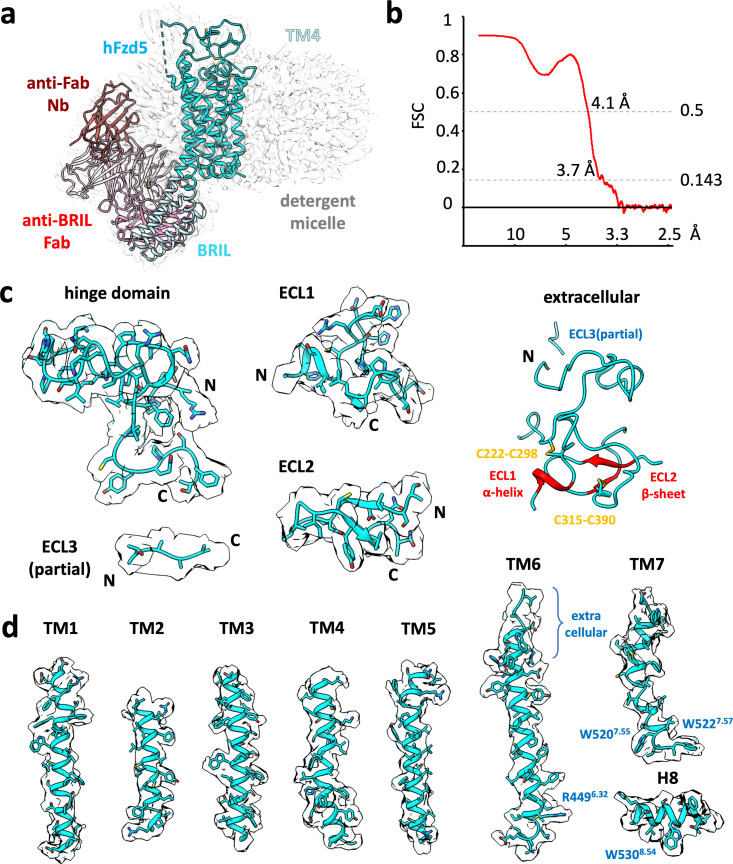
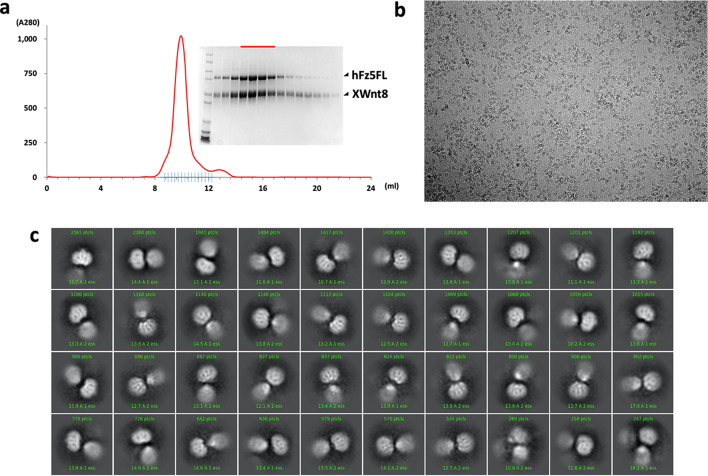
Figure 1. Design scheme and cryo-EM analysis of hFzd5.
(a) Cartoon representation of the strategy for hFzd5 particle decoration by anti-BRIL Fab and anti-Fab Nb. These chaperones double hFzd5 molecular weight and render it asymmetric in detergent micelle, thus working as a fiducial maker for image alignment. (b) A selected 2D class average showing the side view of monomeric hFzd5ICL3BRIL/Fab/Nb. (c) Overall EM volume around the structural model of hFzd5 (cyan), BRIL and the linker between BRIL and Fzd5 (light blue), Fab heavy chain (pink), Fab light chain (light pink) and Nb (wine). (d) The size-exclusion chromatography profile, SDS-PAGE of peak fractions, and (e) the selected 2D class average of XWnt8/hFzd5FL. The full SDS-PAGE and 2D classes are shown in Figure 1—figure supplement 5. Models of XWnt8/mouse Fzd8CRD (mFzd8CRD) (PDB ID: 4F0A) and hFzd5 are overlaid on the blobs to show the relative size of densities.