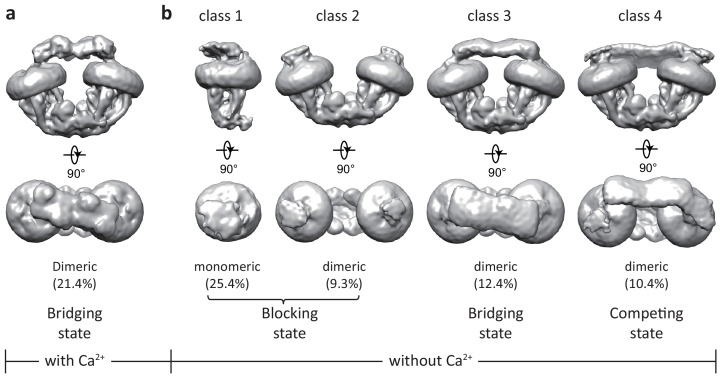
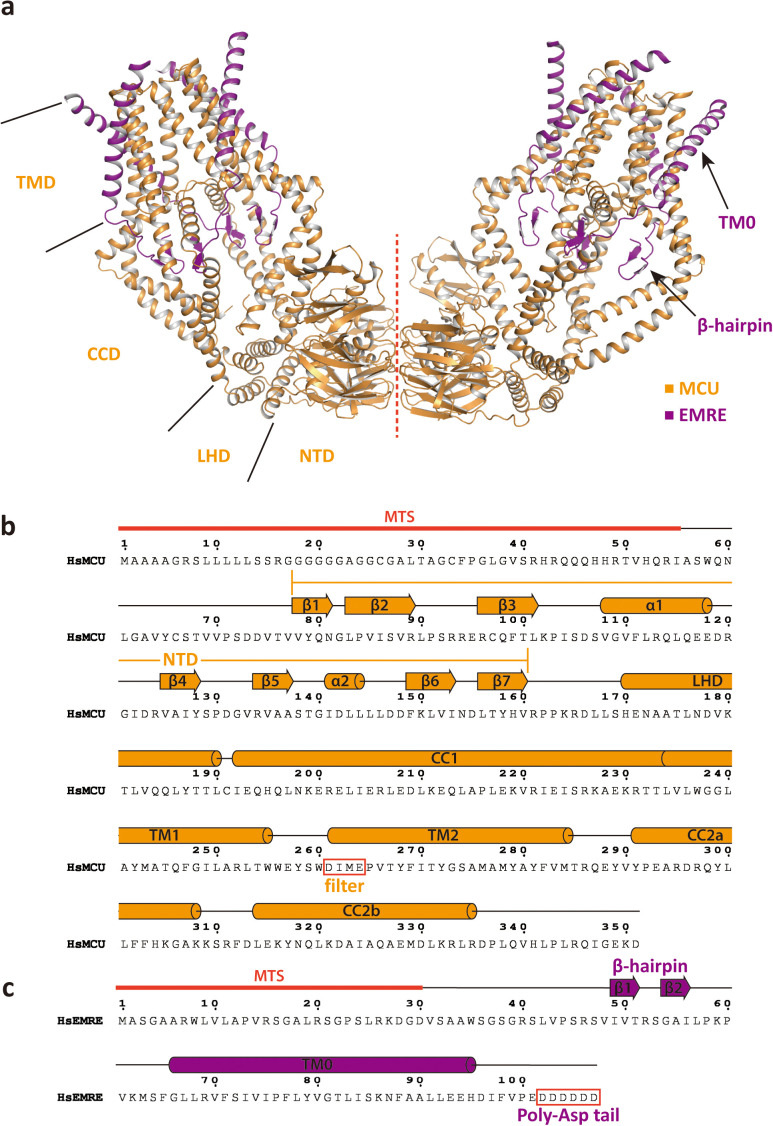
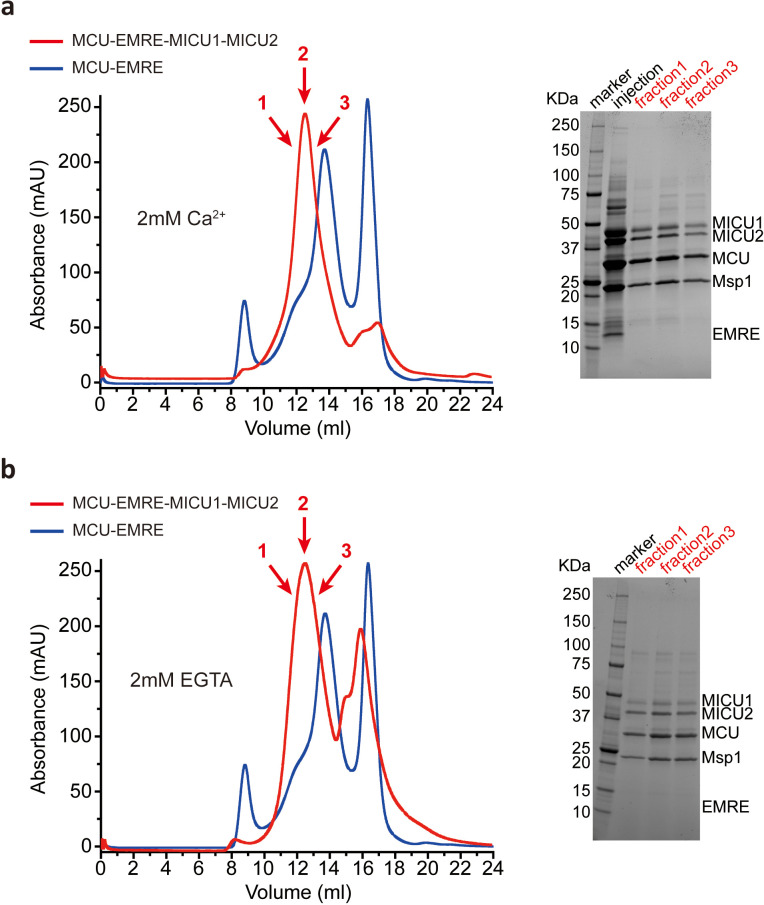
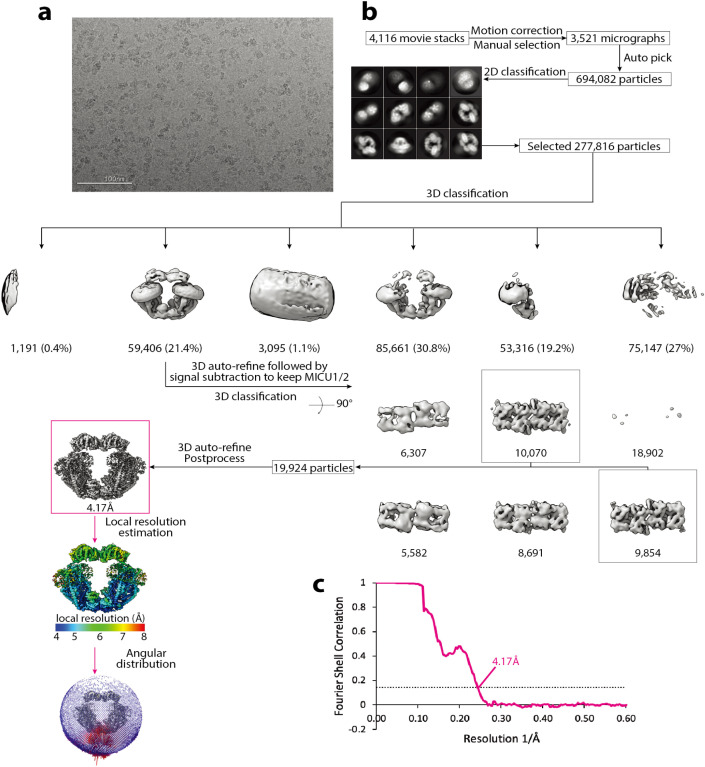
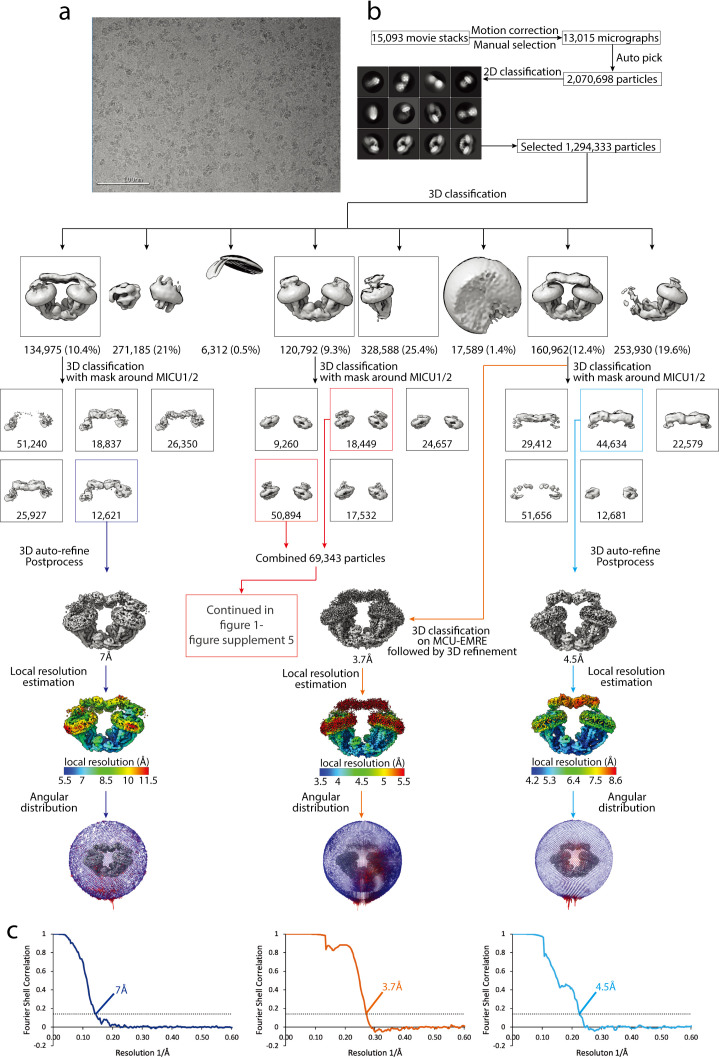
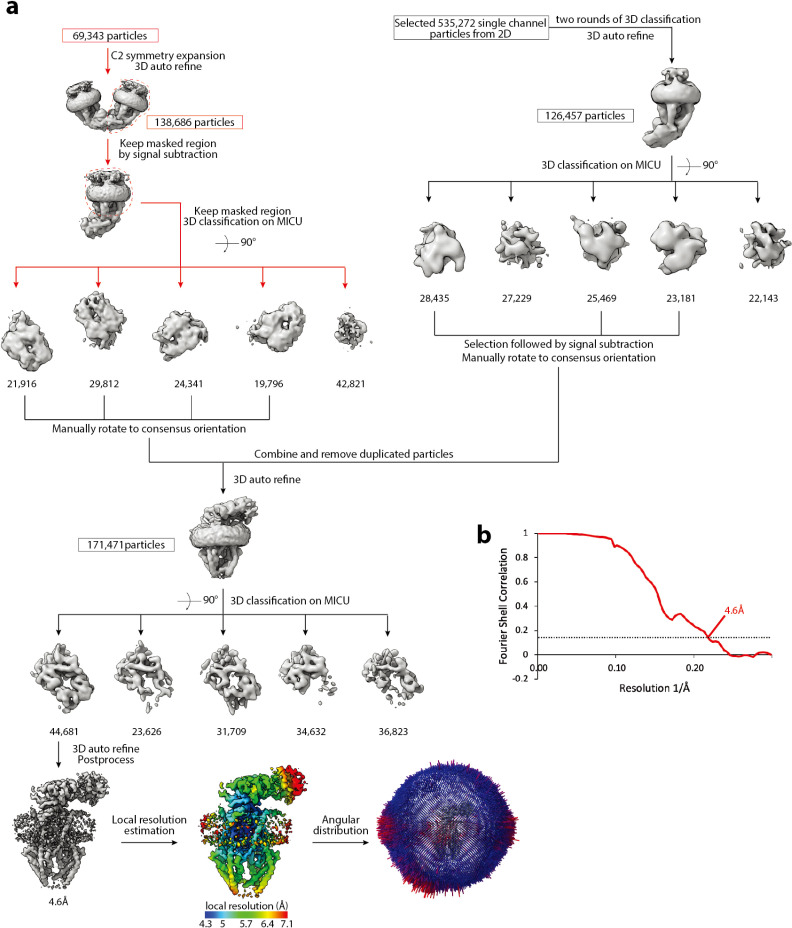
Figure 1. Initial 3D classifications of the human MCU-EMRE-MICU1-MICU2 holocomplex (uniplex) in the presence and absence of Ca2+.
Numbers in parentheses denote the percentage of particles for each class. (a) The major class of particles from the uniplex sample prepared in the presence of 2 mM Ca2+. (b) Four classes of particles from the uniplex sample prepared in the absence of Ca2+ (with 2 mM EGTA).