Figure 5. Key non-catalytic residues regulate LYPLAL1 activity and pharmacological activator efficacy.
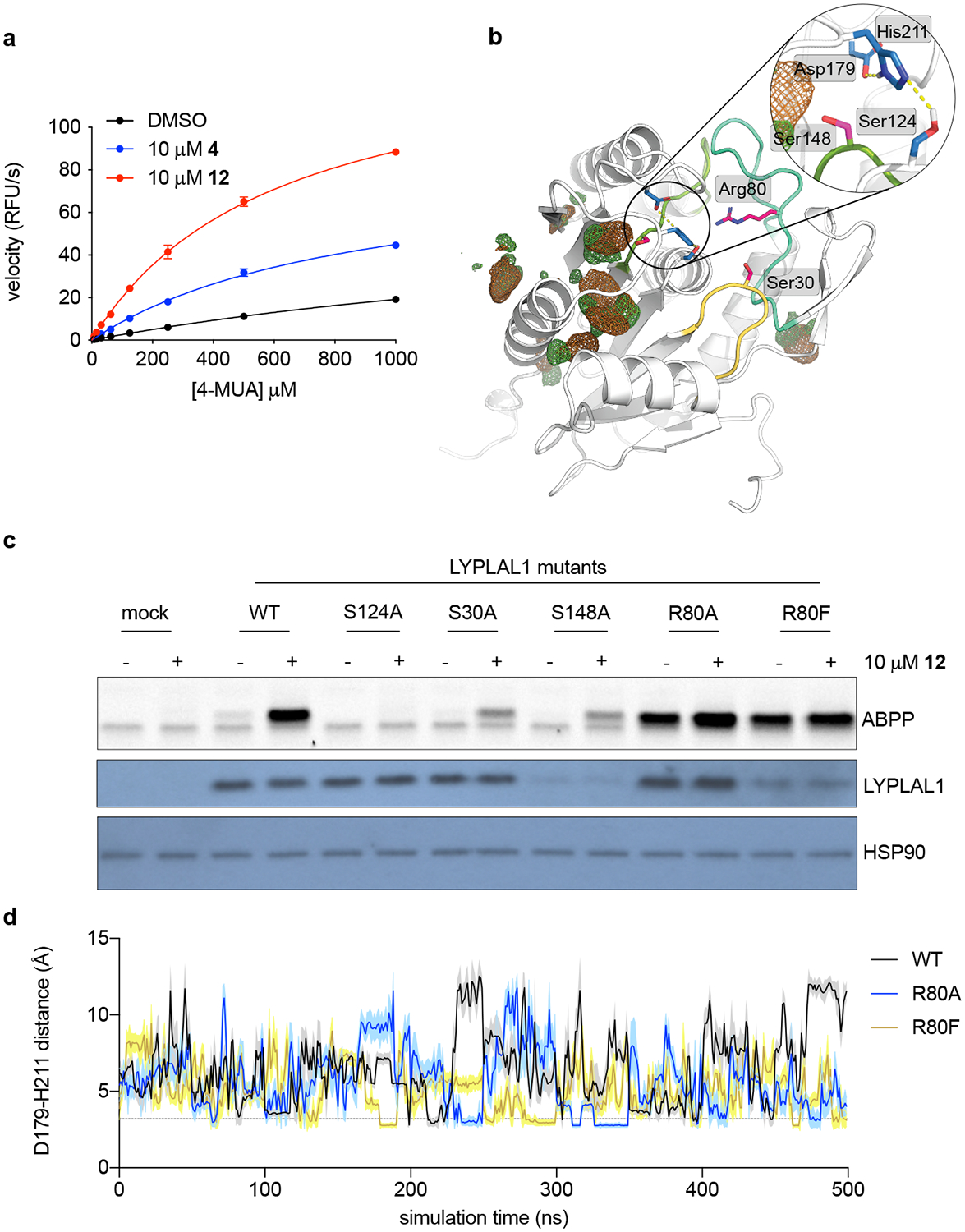
(a) Michaelis-Menten analysis of purified mLYPLAL1 (1 μM) and the synthetic substrate 4-methylumbelliferyl acetate (4-MUA) as shows that 4 and 12 improve catalytic efficiency (n=3 per group representing independent samples). Error bars represent mean ± s.d. (b) Model of LYPLAL1 (PDB 3U0V) highlighting three key residues (Ser30, Arg80 and Ser148 on loops colored yellow, turquoise, and light green) capable of electrostatically interacting with the catalytic triad (residues highlighted in blue). Hydrogen bond interactions within the catalytic triad are shown as yellow dotted lines. Chlorobenzene co-solvent occupancy maps (contoured at 20 σ) are color-coded to represent the different chemical features of the probe used: chlorine (green) and benzene (brown). (c) LYPLAL1 point mutants were expressed in HEK293 cells, proteomes harvested and incubated with vehicle or 12 prior to gel-based ABPP analysis. S124A replaces the catalytic serine and renders the enzyme inactive. Bottom panels show Western blots of LYPLAL1 and HSP90 (loading control). (d) Distance fluctuations between Asp179 and His211 of the catalytic triad during MD simulations of WT LYPLAL1 and Arg80 mutants. Line and shaded region represent mean and s.d. of the intervening data points per ns, respectively. Representative results from three (a, c) independent experiments; similar results were obtained in all experiments. Uncropped gels/blots for c are shown in Supplementary Fig. 6.
