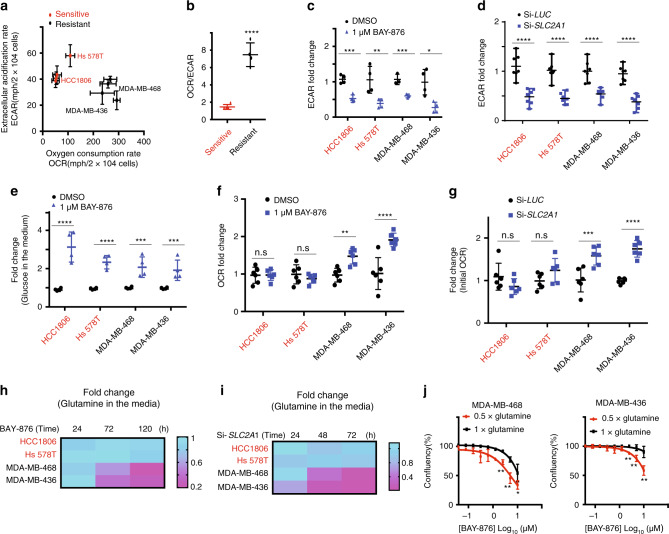
Fig. 2. OXPHOS levels correlate with the response to GLUT1 inhibition.
a OCR and ECAR were measured for each of BAY-876-sensitive lines (red) and resistant lines (black). b OCR and ECAR ratio were calculated for each cell line. Student’s t test, ****p < 0.0001. c ECAR values measurement of cells with or without BAY-876 treatment for 5 days. n = 4; mean ± s.d.; Two-way ANOVA. *p < 0.05; **p < 0.01; ***p < 0.001. d ECAR values measurement of cells with siRNA of GLUT1 knockdown or luciferase control. n = 7; mean ± s.d. Two-way ANOVA. ****p < 0.0001. e Glucose uptake analysis were performed in cells following BAY-876 treatment for the 5 days. n = 4; mean ± s.d. Two-way ANOVA. ***p < 0.001; ****p < 0.0001; f A trace of OCR values from a mitochondrial stress test of cells with or without BAY-876 treatment for 5 days. n = 4; mean ± s.d. Two-way ANOVA. ****p < 0.0001; n.s. not significant. g A trace of OCR values from a mitochondrial stress test of cells with siRNA of GLUT1 or luciferase control. n = 6; mean ± s.d. Two-way ANOVA. ***p < 0.001; ****p < 0.0001; n.s. not significant. h Glutamine uptake analysis was performed in cells following 1 μM BAY-876 treatment. n = 3. i Glutamine uptake analysis performed in cells transfected with 25 nM siSLC2A1 (bottom). n = 2. j Growth curves of MDA-MB-468 cells (left) and MDA-MB-436 cells (right) cultured in complete DMEM medium with or without glutamine deprivation treated with indicated nine doses of BAY-876 for 5 days. n = 3; mean ± s.d. Two-way ANOVA. **p < 0.01; ***p < 0.001; ****p < 0.0001. Source data are provided as a Source Data file.