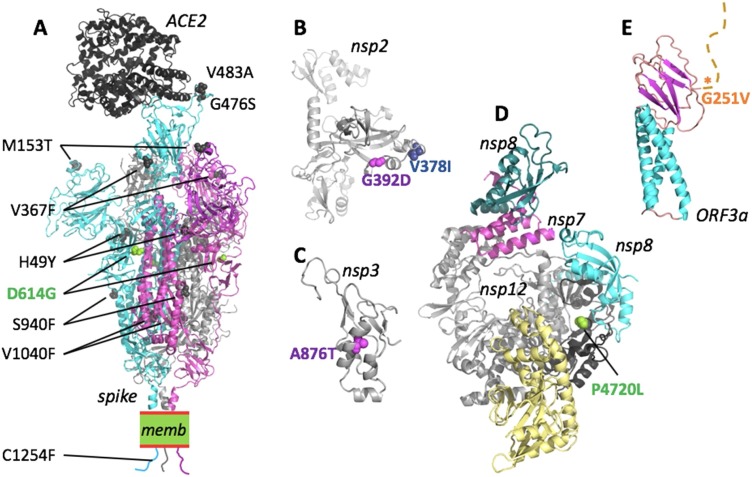
Fig. 4.
Mapping of SARS-CoV-2 clade-defining mutations onto the proteins. Nonsynonymous mutations for proteins where the 3D structure was experimentally determined (spike, nsp12/7/8) or can be inferred with reasonable confidence. Mutations are colour-coded as for the corresponding clades in Fig.3(D: magenta; G: light green; I: blue; V: orange). For a detailed analysis, see Fig.S5-11. (A) The structure of the SARS-CoV-2 spike trimer in its open conformation (chains are cyan, magenta and grey) bound to the human receptor ACE2 (black) modeled based on PDB accessions 6m17 and 6vyb. Identified nonsynonymous mutations are shown as spheres in the model. For reasons of visibility only mutations of two of the three spike chains are labeled. memb. indicates the plasma membrane. (B) Fragment comprising residues 180-534 of nsp2, modelled by AlphaFold35. Both clade-defining mutations are located in solvent-exposed regions and would not lead to steric clashes. (C) The substitution A876 T (corresponding to residue A58 in the nsp3 cleavage product numbering) is situated in the N-terminal ubiquitin-like domain of nsp3. The structure of this domain can be inferred based on the 79% identical structure of residues 1-112 from SARS-CoV (PDB id 2idy). The substitution A876 T can be accommodated with only minor structural adjustments and is not expected to have a substantial influence on the protein stability or function. (D) The structure shows the nsp12 in complex with nsp7 (magenta) and nsp8 (cyan and teal), based on PDB 7btf. P4720 (P323 in nsp12 numbering) is located in the ‘interface domain’ (black). In this position, the P323 L substitution is not predicted to disrupt the folding or protein interactions and hence is not expected to have strong effects. (E) A theoretical model for the Orf3a monomer has been proposed by AlphaFold36. The structure-function relationship of this protein remains to be clarified. The mutation G251 V is located C-terminal to the β-sandwich domain and the tail (marked by an asterisk).