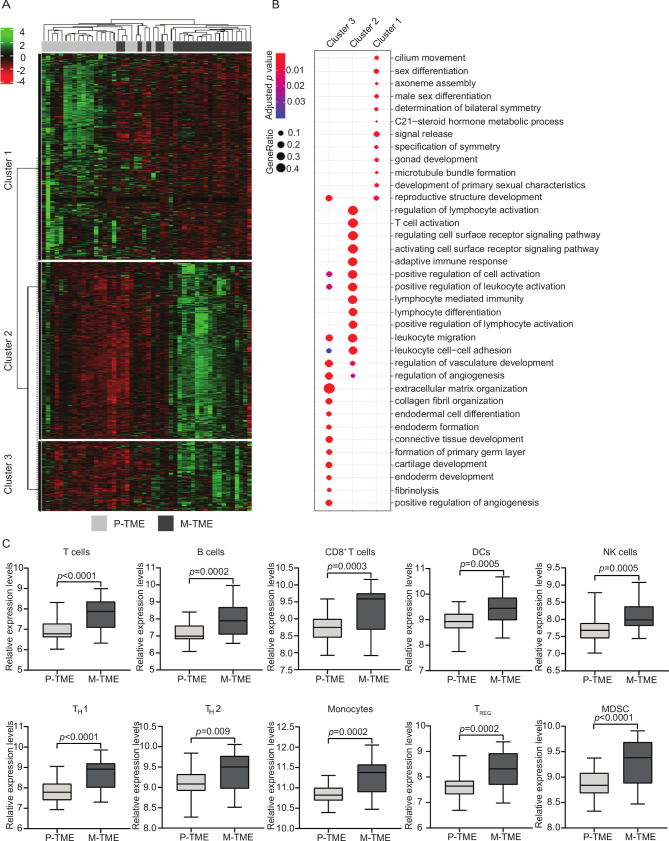
Figure 1.
Transcriptional characterization of primary versus metastatic high-grade serous carcinomas (HGSCs). (A) Hierarchical clustering of 1468 transcripts (fold-change >2, adjusted p-value <0.05) that were significantly changed in paired 24 metastatic tumor microenvironment (M-TME) versus 24 primary tumor microenvironment (P-TME) samples from study group 1, as determined by RNA sequencing. (B) Gene Ontology (biological processes) enrichment analysis of differentially expressed in M-TME versus P-TME. See also online supplementary table 3. (C) Relative expression levels of gene sets associated with T cells, B cells, CD8+ T cells, dendritic cells (DCs), natural killer (NK) cells, TH1 cells, TH2 cells, monocytes, regulatory T (TREG) cells, and myeloid-derived suppressor cells (MDSCs) across paired 24 P-TME and 24 M-TME samples, as determined by metagenes on RNA sequencing data from study group 1. Box plots: lower quartile, median, upper quartile; whiskers, minimum, maximum.