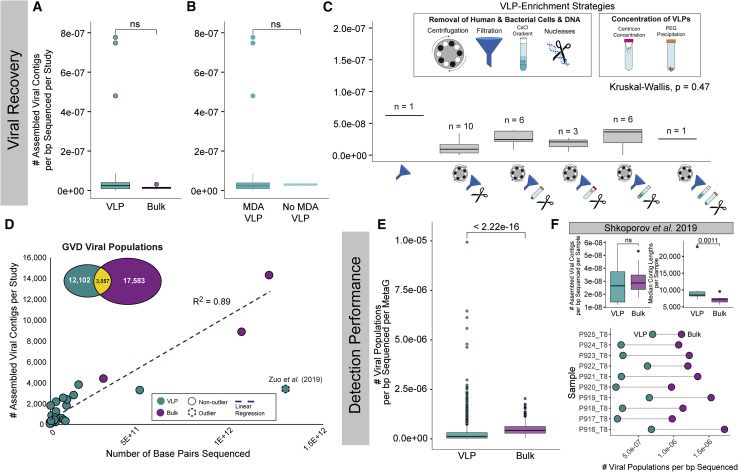
Figure 5.
VLP-Enriched (VLP) and Bulk Metagenomes Comparisons for Studying Viruses in the Human Gut
(A–C) Boxplots showing median and quartiles of the number of assembled contigs per base pair sequenced per study (A) of VLP and bulk metagenomes, (B) of VLP metagenomes with and without MDA, and (C) of the different VLP-enrichment methodologies across the studies. Outlier dots were removed from plot (C) to better show the range of values. The n value above each box plot represents the number of studies using each VLP-enrichment method.
(D) Scatter plot with a linear regression line showing the number of assembled viral contigs per bp sequenced per study with VLP and bulk metagenome studies identified by different colors. In the inset is a Venn diagram showing the number of GVD viral populations that originated from VLP or bulk or both types of metagenomes.
(E) Boxplots showing median and quartiles of the number of viral populations detected per bp sequenced per individual of VLP and bulk metagenomes.
(F) Boxplots showing median and quartiles of the number of assembled contigs per bp sequenced (top left) and the median contig length (top right) for VLP and bulk metagenomes processed for the same samples in the Shkoporov et al, (2019) (bottom). Connected dot plot showing the number of viral populations detected per bp sequencedby using VLP and bulk metagenomes for each individual in the Shkoporov et al, (2019) study. All pairwise comparisons were performed by using Mann-Whitney U tests. Non-significant p values are denoted as “ns.”
See also Figure S4.