Figure 2.
Input Specificity to Dopaminergic Neurons Matches Anatomical Subtypes
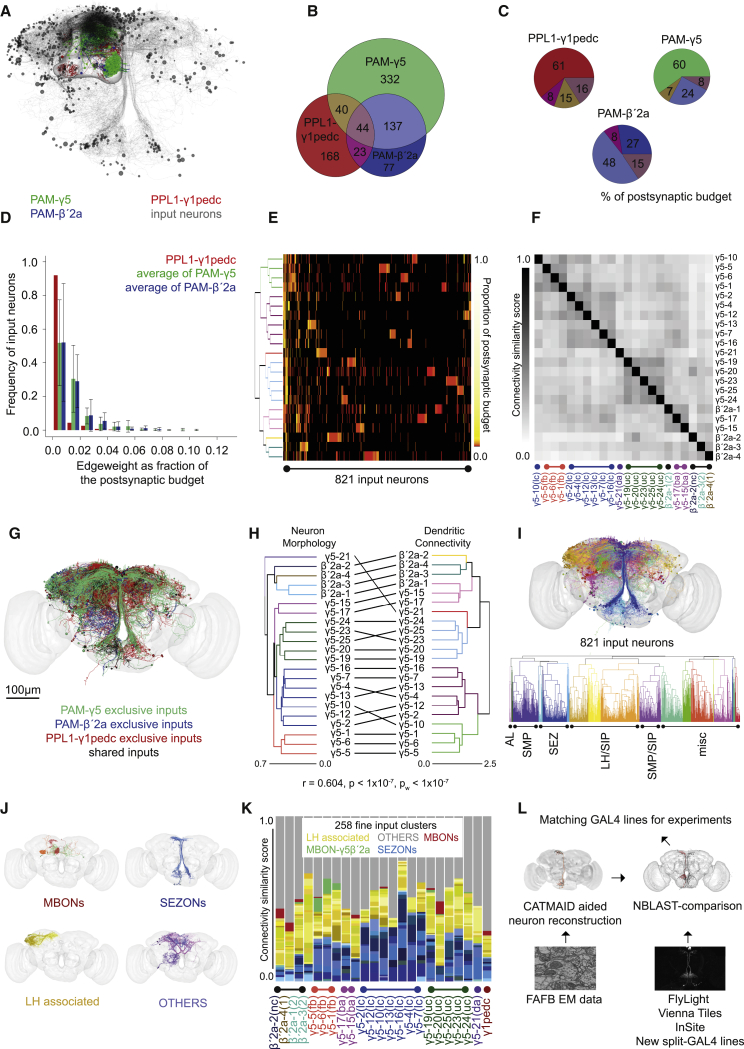
(A) Representation of all 821 input neurons to PAM-γ5, PAM-β′2a, and PPL1-γ1pedc DANs identified in this study. Cell bodies (black spheres) and processes (gray). DANs and the MB outline shown for reference (compare to Figure 1A).
(B) Venn diagram of unique and common input neurons to the analyzed DANs. PPL1-γ1pedc receives largely different input to PAM-γ5 and PAM-β′2a DANs. PAM-γ5 and PAM-β′2a DANs have many common inputs and some are also shared by PPL1-γ1pedc.
(C) Pie charts showing percentage of postsynaptic budget occupied by shared and unique input neurons to PAM-γ5, PAM-β′2a, and PPL1-γ1pedc DANs. Percentage of shared inputs across all three groups is 16%, 15%, and 8% for PPL1-γ1pedc, PAM-β′2a, and PAM-γ5 DANs, respectively.
(D) Bar chart showing DANs have many inputs with very low edge weight and each representing a small fraction of their overall postsynaptic budget. Inputs contributing more of the postsynaptic budget (to the right of the graph) are more abundant for PAM-γ5 and PAM-β′2a DANs; PPL1-γ1pedc distribution is strongly left shifted (bars show mean ± SD).
(E) DANs can be clustered by input connectivity (rows correspond to F). Heatmap shows every DAN has a group of unique input neurons represented by unique blocks in each row. Clustering of DANs mostly depends on lesser number of shared inputs compressed to the left edge of the heatmap.
(F) A matrix where DANs are grouped by the similarity of their input connectivity has clear structure, i.e., significantly more organized than random connectivity (comparison to null model, p < 0.0001; see Methods S1).
(G) Representation of traced input neurons labeled using the unique and common input anatomy determined in (B) (see also Figures S2A–S2D).
(H) Tanglegram comparing DAN clustering by morphology (from Figure 1B) and clustering by input connectivity (left of E). Connectivity and morphology are not significantly independent of each other (Pearson’s correlation between the corresponding distance matrices, r = 0.604; Mantel test, p < 10−7; pw < 10−7 within only γ5 or β′2a group).
(I) DAN input neurons clustered by morphology. Dendrogram below shows single neurons allocated to 20 major coarse clusters based on soma position and primary neurite tract. Approximate neuropil of origin is indicated: antennal lobe (AL), SMP, SEZ, LH/SIP, and SMP/SIP are marked. Many neurons originate from less explored neuropils (misc).
(J) Fine clusters of exemplary neurons for the MBON, LHON, SEZON, and OTHERS classes of DAN inputs.
(K) Bar plot showing respective number of MBON, LHON, SEZON, and OTHERS inputs to individual DANs, ordered according to cluster identity (Figure 1). In general, PAM-γ5 DANs receive about 35% of their input from SEZONs, and about 20% from LHONs. Only the 3 PAM-γ5(fb) DANs receive significant direct input from MBON-γ5β′2a (green segments). PAM-β′2a DANs receive about 15% from SEZONs and 35% from LHONs. One PAM-β′2a DAN also receives minor direct input from MBON-γ5β′2a. PPL1-γ1pedc DANs receive roughly equal LHON and SEZON input.
(L) NBLAST compares CATMAID generated neuronal skeletons from FAFB to neurons labeled in confocal images of GAL4 expression patterns.
See also Figure S2, Methods S1, and Video S3.