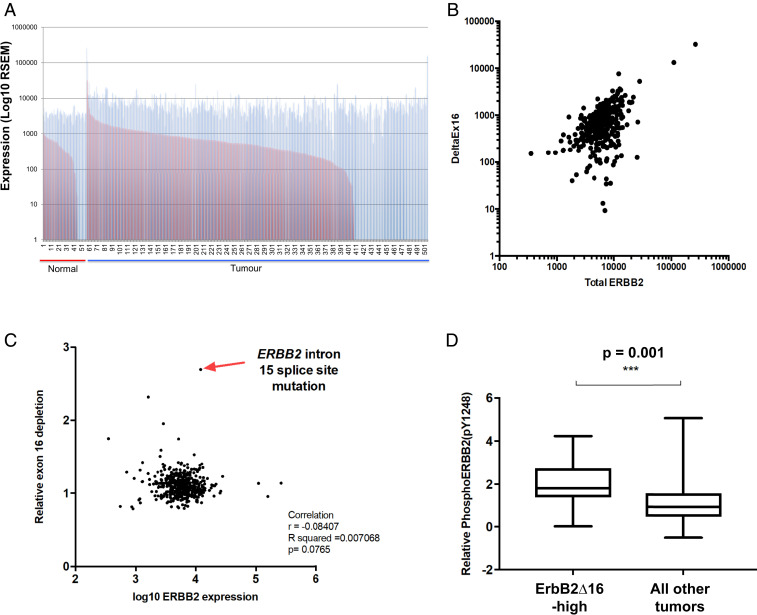
Fig. 1.
A subset of human lung cancers with elevated expression of ErbB2ΔEx16 and tyrosine phosphorylation of ERBB2. (A) RNA-seq data from the TCGA lung adenocarcinoma dataset were analyzed to quantify full-length ERBB2 (blue bars) and ERBB2ΔEx16 (red bars) transcripts in samples of normal lung tissue and lung cancers. (B) The relationship between overall ERBB2 transcript level and ERBB2ΔEx16 level in the TCGA lung adenocarcinoma RNA-seq data was examined using Pearson’s correlation analysis. (C) The ratio of ERBB2ΔEx16 to full-length ERBB2 was plotted against the total level of ERBB2 transcript as determined from TCGA lung adenocarcinoma RNA-seq data, while identifying the tumor sample with highest ERBB2ΔEx16:ERBB2 ratio (arrow), which harbors a point mutation ablating the splice acceptor site in intron 15. (D) TCGA RPPA data for ERBB2 tyrosine 1248 phosphorylation in a subset of the same panel of lung adenocarcinoma samples was stratified by expression level of ERBB2ΔEx16 whereby the 5% of tumors with the highest ERBB2ΔEx16 transcript levels were considered ERBB2ΔEx16-high.