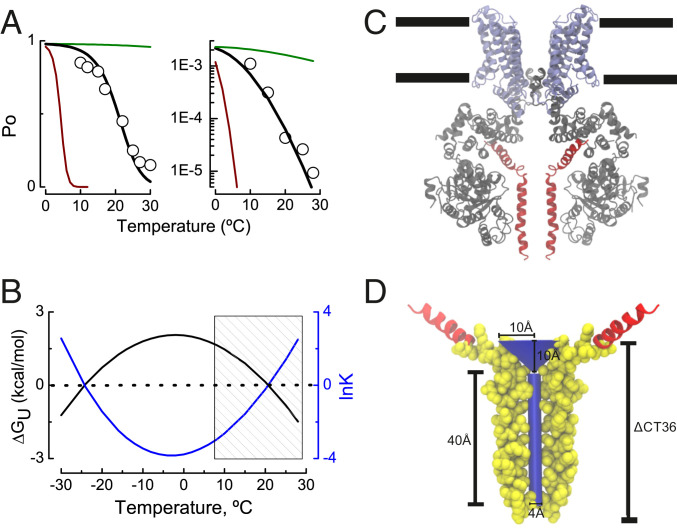
Fig. 4.
A thermodynamically inspired–structure-supported model for TRPM8 channel gating. (A) TRPM8 channel open probability recorded at +260 mV (Left) and the minimal channel open probability recorded at very negative potentials using the limiting slope method (Right) at temperatures ranging from 10 to 30 °C according to ref. 32 (hollow circles). Black line represents the fit of Eqs. 14 and 15 to the experimental data from Raddatz et al. (32). Best fit parameters were ΔCp = 2.2 kcal/mol*K and ΔSo = −7.6 cal/molK (Left) and ΔCp = 2.2 kcal/mol*K and ΔSo = 12 cal/mol*K (Right). To = 270K (−3 °C) in both panels. The green and red lines in Fig. 1A are the Po values between 0 and 30 °C according to Eqs. 14 and 15 using ΔCp = 0.22 and 22 kcal/mol*K, respectively. (B) Difference in the free energy of unfolding ΔGU (kcal/mol) between resting–active state of the TRPM8 temperature sensor according to Eqs. 4 and 11 (fit parameters as in Fig. 4A, Left). Shaded area depicts the temperature range where the experiments in ref. 32 were performed. (C) Ligand-free TRPM8 structure obtained by spCryo-EM (PDB ID 6O6A; ref. 9), where the N terminus domain is colored in gray, transmembrane segments 1 to 6 and TRP domain appear in purple, and distal CTD is in red (subunits in the front and in the back were removed for clarity). Black lines define the approximate lipid bilayer limits. (D) The deleted CTD section (ΔCT36) allocates a 40 Å × 2 Å (length × radius) cylinder of volume 503 Å3 plus a cone of 10 Å × 10 Å (radius × height), with a volume of 1,047 Å3. The segment encompassing the 36 amino acids whose deletion decreases the channel temperature dependence (TRPM8ΔCT36) is depicted in Van der Waals representation using the Visual Molecular Dynamics (VMD) software (66).