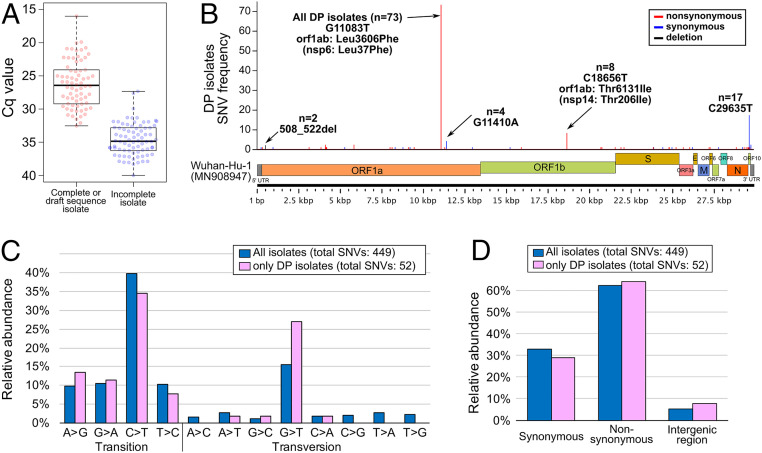
Fig. 1.
WGS of SARS-CoV-2 isolates (strains) on the DP cruise ship off Yokohama in Japan. (A) Box plots of the Cq values from RT-qPCR assays that either successfully resulted in WGS or incomplete sequencing. A Cq of at least 32 (∼300 viral copy) is favorable for obtaining a whole-genome sequence by PrimalSeq. (B) The genome position of genetic variations found among DP isolates compared with the isolate Wuhan-Hu-1 (MN908947). (C) The detected nucleotide transition and transversion in all GISAID available SARS-CoV-2 genomes (n = 412, isolation date by February 19, 2020; updated on May 14, 2020) (Dataset S3) and DP isolates (n = 73) compared with the isolate Wuhan-Hu-1 (MN908947). (D) The detected ratio of synonymous/nonsynonymous mutations on coding sequences or intergenic regions in all GISAID available SARS-CoV-2 genomes is described above (n = 412) and DP isolates (n = 73) compared with the isolate Wuhan-Hu-1 (MN908947).