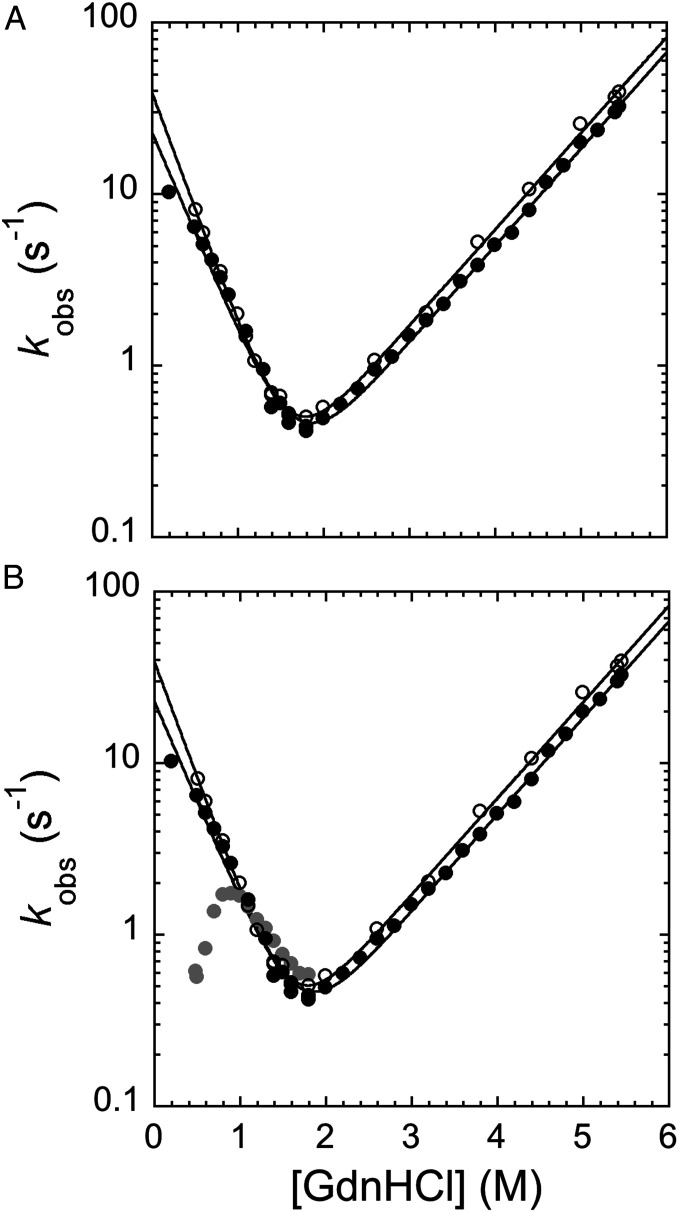
Fig. 3.
Unfolding and refolding kinetics of pP1-P2 and pPDZ1. (A) Chevron plot of pPDZ1 (empty circles) and pP1-P2 (black circles) obtained when starting the refolding experiment by diluting the protein with mild denaturant concentrations (i.e., 2.2 M GdnHCl). The two chevron plots are perfectly superimposable, indicating that in this condition, the folding of PDZ1 is not affected by the presence of a flanking domain when PDZ2 is held in its native conformation (see the text for details). (B) Chevron plot of pPDZ1 (empty circles) and pP1-P2 obtained when starting the refolding by diluting the protein with a mild denaturant concentration (i.e., 2.2 M GdnHCl; black circles) and a high denaturant concentration (i.e., 5.37 M GdnHCl; gray circles). As explained in detail in the text, when the refolding starts at higher GdnHCl concentrations, both PDZ1 and PDZ2 domains are denatured, leading to accumulation of a kinetic trap (represented by the presence of a pronounced rollover in the refolding arm) that competes with the productive folding of the protein. The buffer used in both experiments was 50 mM Tris⋅HCl pH 7.5 and 0.3 M NaCl.