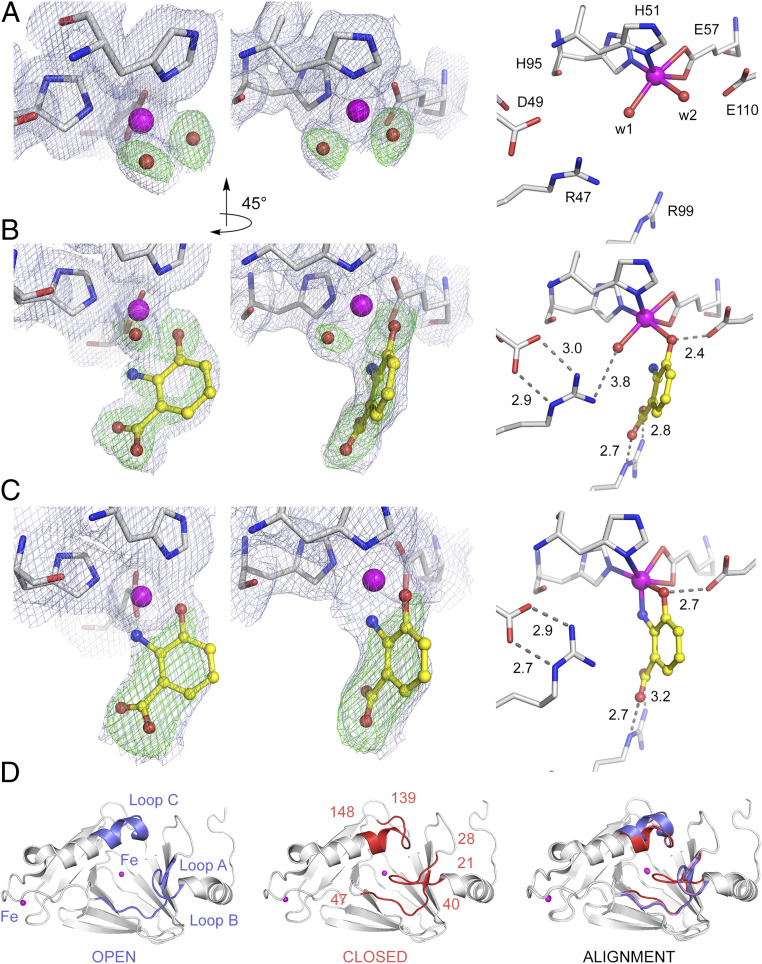
Fig. 3.
A two-step 3-HAA binding process. (A) The ligand-free structure in resting enzyme, (B) the transient (E•S)I intermediate after ∼20-s exposure to 3-HAA, and (C) the equilibrium (E•S)II complex observed after incubation periods of 30 s with 3-HAA. The light blue 2Fobs – Fcalc maps and green omit Fobs – Fcalc maps are contoured at 1.0 σ and 3.0 σ, respectively. Atom color code: gray, carbon (protein residues); yellow, carbon (ligand); blue, nitrogen; red, oxygen; magenta, iron. (D) The crystal structure of the substrate-free enzyme is in a fully open form (blue), and the transient monodentate (E•S)I intermediate exhibits a partially closed conformation (pink), whereas the structure of the bidentate (E•S)II complex is in a fully closed conformation (red). Significant loop flexibility is observed at three specific loop regions consisting of highly conserved residues 21 to 28 of loop A, 40 to 47 of loop B, and 139 to 148 of loop C.