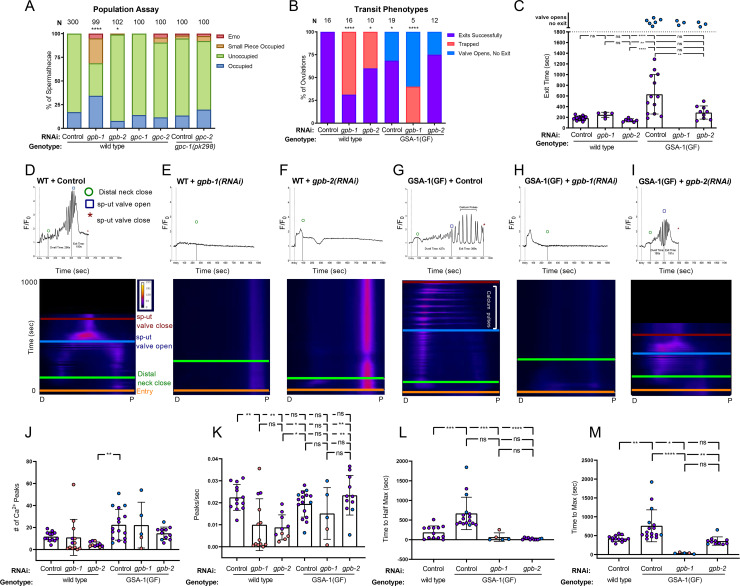
Fig 6. Heterotrimeric G-protein beta subunits GPB-1 and GPB-2 regulate Ca2+ signaling and spermatheca contractility.
(A) Population assay of wild type young adults grown on control RNAi, gpb-1(RNAi), gpb-2(RNAi), gpc-1(RNAi), and gpc-2(RNAi) and gpc-1(pk298) grown on control RNAi and gpc-2(RNAi). Spermathecae were scored for the presence or absence of an embryo (occupied or unoccupied), presence of a fragment of an embryo (small piece occupied), or the presence of endomitotic oocytes in the gonad arm (emo) phenotypes. The total number of unoccupied spermatheca was compared to the sum of all other phenotypes using the Fisher’s exact t-test. N is the total number of spermathecae counted. (B) Transit phenotypes of the ovulations in wild type animals treated with control RNAi, gpb-1(RNAi) and gpb-2(RNAi), and GSA-1(GF) animals treated with control RNAi, gpb-1(RNAi), and gpb-2(RNAi) were scored for successful embryo transits through the spermatheca (exits successfully), failure to exit (trapped), and the situation in which the sp-ut valve opens, but the embryo does not exit (valve opens, no exit). Color coding of the data points corresponds to the transit phenotypes in B. For transit phenotype analysis, the total number of oocytes that exited the spermatheca successfully was compared to the sum of all other phenotypes. Fisher’s exact t-tests were used for both population assays and transit phenotype analysis. (C) Exit time of movies in B were compared using One-way ANOVA with a multiple comparison Tukey’s test. Representative normalized Ca2+ traces and kymograms of movies in B (D-I) are shown with time of entry, distal neck closure, and time the sp-ut valve opens and closes indicated. Levels of Ca2+ signal were normalized to 30 frames before oocyte entry. Kymograms generated by averaging over the columns of each movie frame (see methods). Refer to S3A, S3E, S3F Fig and S5A, S4C, S4D Figs for additional Ca2+ traces, and S6D and S6E Fig and S9B and S9C Fig for additional kymograms. (J-M) Color coding of the data points corresponds to the transit phenotypes in A. (J) The number of Ca2+ peaks and (K) the peaks per second was determined for wild type animals treated with control RNAi, gpb-1(RNAi) and gpb-2(RNAi), and GSA-1(GF) animals treated with control RNAi, gpb-1(RNAi), and gpb-2(RNAi). The amount of time after oocyte entry required to reach either the (L) half maximum or (M) maximum Ca2+ signal was quantified for wild type animals treated with control RNAi, GSA-1(GF) animals treated with control RNAi, gpb-1(RNAi), and gpb-2(RNAi). These values were compared using one-way ANOVA with a multiple comparison Tukey’s test. and compared using Fishers exact t-test. Stars designate statistical significance (**** p<0.0001, *** p<0.005, ** p<0.01, * p<0.05).