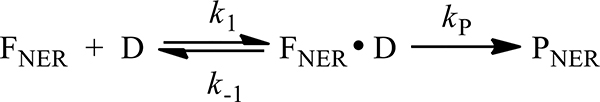Abstract
The excision of DNA lesions by human Nucleotide Excision Repair (NER) has been extensively studied in human cell extracts. Employing DNA duplexes less than 200 base pairs (bp) in lengths containing single bulky, benzo[a]pyrene-derived guanine lesion (B[a]P-dG), the NER yields are typically of the order of ~ 5 – 10%, or less. Remarkably, the NER yield is enhanced by a factor of ~ 6 when the B[a]P-dG lesion is embedded in a covalently closed circular pUC19NN plasmid (contour length 2686 bp) than in the same plasmid linearized by a restriction enzyme with the B[a]P-dG adduct positioned at the 945th nucleotide counted from the 5’-end of the linearized DNA molecules. Furthermore, the NER yield in the circular pUC19NN plasmid is ~ 9 times greater than in a short 147-mer DNA duplex with the B[a]P-dG adduct positioned in the middle. Although the NER factors responsible for these differences were not explicitly identified here, the hypothesis is proposed that the initial DNA damage sensor XPC-RAD23B is a likely candidate; it is known to search for DNA lesions by a constrained one-dimensional search mechanism (Cheon et al., Nucleic Acids Res. 2018) and our results are consistent with the notion that it dissociates more readily from the blunt ends than from the inner regions of linear DNA duplexes, thus accounting for the remarkable enhancement in NER yields associated with the single B[a]P-dG adduct embedded in covalently closed circular plasmids.
Graphical Abstract
INTRODUCTION
The human genome is under continuous attack by highly reactive intermediates produced by ionizing radiation and environmental pollutants that damage the genetic material of human cells.1, 2 In healthy tissues, the integrity of the human genome is maintained by a DNA repair system that removes DNA lesions in an efficient and timely manner.3 Bulky, DNA double helix-distorting lesions are excised by the Nucleotide Excision Repair (NER) mechanism4 yielding dual incision oligonucleotide products 24 – 32 nucleotides in lengths containing the lesion. Optimal NER activity is observed when there are at least 44 base pairs (bp) available on the 3’-side and > 50 on the 5’-side of the lesion.5 Therefore, in vitro NER studies in mammalian cell extracts are usually conducted with DNA duplexes at least 130 – 160 bp in lengths.6, 7 However, it is unclear whether NER efficiencies are affected if the lesions are embedded in even longer double-stranded DNA duplexes, or covalently closed circular DNA plasmids. Another question is the impact of double-stranded DNA termini on NER yields. It has been demonstrated that the DNA end-binding protein Ku70/80 can suppress the NER excision of 1,3-GTG-Pt(II) intrastrand crosslinks embedded in 149 bp DNA duplexes in human cell extracts.8 The binding of the heterodimeric DNA damage sensor XPC-RAD23B (XPC) to the ends of linear DNA molecules with single stranded overhangs is known to be enhanced9 and thus could potentially reduce NER efficiencies as well.
In this work, we compared the relative NER dual incision efficiencies of the site-specifically inserted bulky DNA lesion in a covalently closed plasmid pUC19NN (Figure 1) and the same, but linearized plasmid 2686 bp in length with blunt ends, and also in a fully base-paired 147-mer DNA duplex in human cell extracts. The design of the covalently closed circular plasmid harboring site-specifically positioned DNA lesion is of particular interest, since the putative effects of DNA end-binding proteins are eliminated.
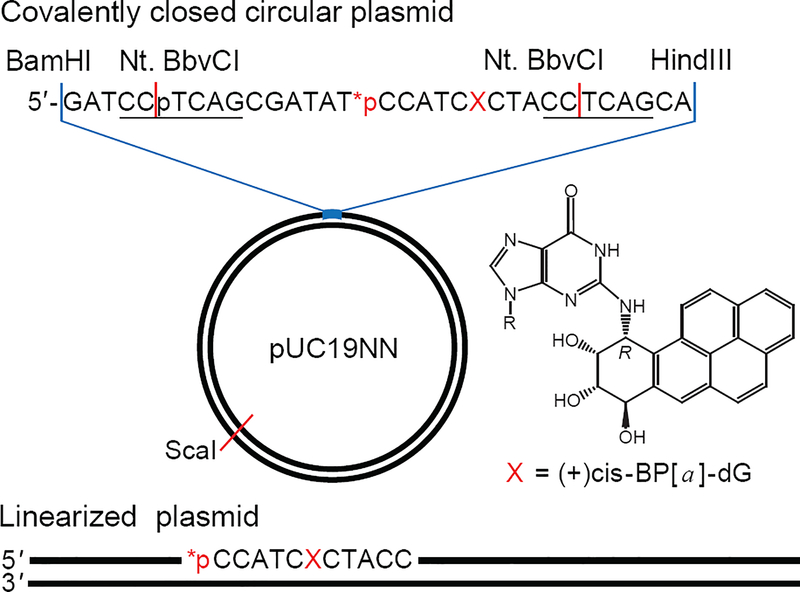
Figure 1.
Covalently closed circular and linearized plasmid substrates (2686 bp) containing site-specifically positioned (+)-cis-B[a]P-dG adduct (X) and a radioactive 32P-internal label. The covalently closed plasmid substrate was generated by an established gapped-vector technology10 from a pUC19NN plasmid containing two Nt. BbvCI restriction sites (underlined). The linearized plasmid substrate with the lesion X positioned at the 945th nucleotide counted from the 5′-end was prepared by the selective cleavage of the circular plasmid with a unique ScaI restriction enzyme that generates blunt end cleavage products.
The site-specifically positioned lesion was the 10R (+)-cis-anti-B[a]PDE-N2-dG adduct (abbreviated as B[a]P-dG), an excellent NER substrate in double-stranded DNA.11, 12 The kinetics of the NER dual incisions of all substrates were monitored in HeLa cell extracts.2
EXPERIMENTAL PROCEDURES
Materials
The plasmid pUC19 and restriction enzymes were obtained from New England Biolabs (Ipswich, MA). The T4-kinase, T4 ligase, and proteinase K were purchased from ThermoFisher Scientific. The 2′-oligodeoxynucleotides were obtained from Integrated DNA Technologies (Coralville, IA) and purified by denaturing polyacrylamide gel electrophoresis before use.
Preparation of pUC19NN Plasmid
The pUC19NN plasmid (Figure 1) was cloned from the pUC19 plasmid. The latter was sequentially treated with HindIII and BamHI restriction enzymes, and the HindIII/BamHI digest was purified by E.Z.N.A. Cycle Pure Kit (Omega Bio-tek, Norcross, GA), annealed to the 5′-phosphrylated insert, and ligated by T4 ligase at 16 °C for 16 h. This insert containing two Nt. BbvCI restriction sites (underlined) was prepared by annealing the 5′-pGATCCTCAGCGATATCCATCGCTACCTCAGCA and 5′-pAGCTTGCTGAGGTAGCGATGGATATCGCTGAG oligonucleotides with non-cohesive overhangs shown in bold. The ligation mixtures were transformed directly into DH5α competent E. coli on LB agar plates, supplemented with 100 mg/L of ampicillin. After screening with Bioworld X-Gal-IPTG, the white colonies were randomly picked, multiplied and isolated with Qiagen Plasmid Mini Kit. Before the large-scale plasmid preparation and isolation with the Plasmid Maxi Kit, incorporation of the insert was confirmed by Sanger sequencing (Genewiz, South Plainfield, NJ). Two G:C base pairs flanking the insert were deleted during the cloning process (Figure S1, Supporting Information). Both deletions were irrelevant to the function of pUC19NN as they did not affect the Nt. BbvCI restriction sites. The integrity of the covalently closed circular pUC19NN was confirmed by 1% agarose gel electrophoresis.
Construction of Plasmid Substrates Harboring single B[a]P-dG Adducts
The plasmid substrates containing site-specifically positioned lesions were generated from pUC19NN plasmids employing a gapped vector technology.10 Aliquots (50 μL) of pUC19NN (5 pmol) were treated with Nt. BbvCI restriction enzyme (50 U) at 37 °C for 1.5 h. The double-nicked pUC19NN was mixed with an excess of the complementary strand 5′-GGTAGCGATGGATATCGCTGA (500 pmol), heated to 80 °C for 5 min and cooled on ice for 2 min (this cycle was repeated 3 times). The gapped plasmid purified by diafiltration on Amicon Ultra-0.5 spin column (100 kDa cut-off) was annealed to the 5′-phosphorylated oligonucleotides, 5′-pTCAGCGATAT and radiolabeled 5′−32pCCATCXCTACC containing the lesion (X = B[a]P-dG) and ligated by T4 ligase at 16 °C for 16 h. The reaction products were treated by T5 Endonuclease to generate linear and nicked plasmids.13 The covalently closed circular plasmids were purified by diafiltration on Amicon Ultra-0.5 spin columns (100 kDa cut-off). The integrity of the circular plasmid substrates was confirmed by 1% agarose gel electrophoresis using both 32P and fluorescence (Ethidium Bromide) detection.
Construction of 147 bp DNA Substrates
The 147 bp DNA duplexes were equivalent to the Widom 601 strong positioning DNA sequence,14 except for the 11-mer 5’-CCATCXCTACC (X = B[a]P-dG) fragment (Figure 1). This 5′−32P-labeled 11-mer was inserted into 147-mer sequences by replacing an equal number of 601 sequence nucleotides and the ends were ligated by the phosphorylation/ligation synthesis methods; the lesions were positioned at the 66th nucleotide counted from the 5’-end of the 147-mer strand.15 The 147-mer modified sequences obtained by this approach were purified by denaturing PAGE and annealed to fully complementary strands bearing C-bases opposite the lesion.
Linearized plasmid substrates
The covalently closed circular plasmids were treated with the ScaI restriction enzyme (Figure 1). The non-circular nature of the plasmid substrates was confirmed by 1% agarose gel electrophoresis.
DNA Repair Assays in HeLa Cell Extracts
NER-competent whole-cell extracts were prepared from HeLaS3 cells (Cell Culture Company, Minneapolis, MN) as described in detail elsewhere.16, 17 The extracts in NER-dialysis buffer (25 mM HEPES, pH 7.9, 70 mM KCl, 12 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 12% glycerol; protein concentration: 12 – 14 mg/mL by Bradford assay) were aliquoted, frozen in liquid nitrogen, and stored at – 80 °C. The NER dual incision reactions were performed in the reaction solutions (total volume 100 μL) containing 25 mM HEPES, pH 7.9, 70 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 4 mM ATP, 200 μg/μL bovine serum albumin, 4% glycerol, 20 fmol of 32P-labeled DNA substrate and 30 μL HeLa cell extract. After incubation at 30 °C for fixed period of time the reactions were stopped by the addition of 40 μg proteinase K in 0.3% SDS and incubated for 30 min at 37 °C. After phenol/chloroform extraction, the low molecular weight DNA was ethanol precipitated and subjected to denaturing 12% polyacrylamide gel electrophoresis.
Analysis of the Gel Images
The polyacrylamide gels were dried, exposed to Molecular Dynamics Storage Phosphor Screens, and then scanned with a Typhoon FLA 9000 laser scanner. The gel autoradiographs were analysed using the Total Lab TL120 1D software.
RESULTS AND DISCUSSION
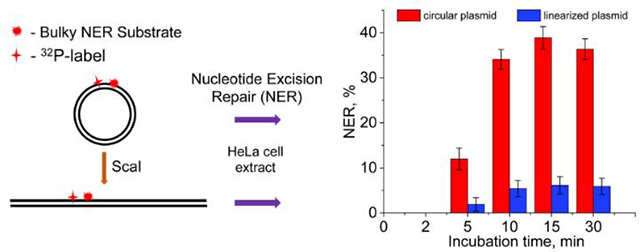
A hallmark of successful NER activity is the appearance of the oligonucleotide dual incision products of ~24 – 32 nucleotides in lengths.18, 19 In order to explore the impact of DNA-end effects and the lengths of DNA substrates on the efficiencies of NER dual incisions in human cell extracts, we constructed plasmid substrates that contained a single site-specifically inserted bulky DNA lesions (Figure 1). The (+)-cis-B[a]P-dG adduct (X) is a well-known NER substrate generated by the exposure of DNA in aqueous solutions to the highly reactive metabolite of the environmental pollutant and carcinogen benzo[a]pyrene,20 (+)-7R,8S-dihydrodiol, 9S,10R-epoxy-tetrahydrobenzo[a]pyrene enantiomer (B[a]PDE). This stereoisomeric bulky adduct is ideal for studying difference in NER incision efficiencies in human cell extracts.11, 12 The cleavage of the covalently closed circular plasmid substrate by a unique restriction enzyme ScaI generates a long linearized plasmid substrate with blunt ends and the B[a]P-dG adduct positioned at the 945th nucleotide counted from the 5′-end.
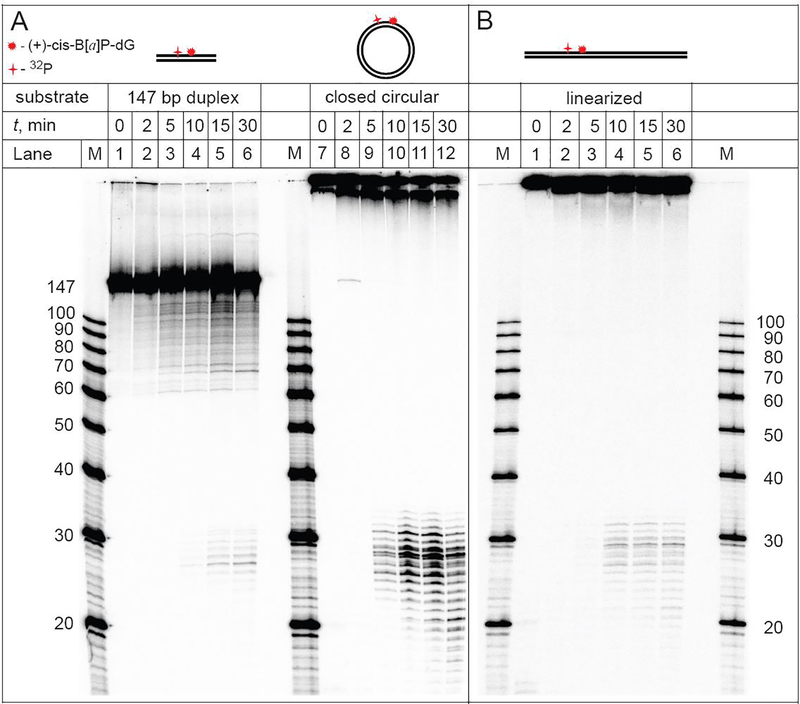
Typical results of the DNA repair assays in HeLa cell extracts obtained with the 32P-internally labeled covalently closed circular plasmid, linearized plasmid, and control linear 147 bp duplexes in equal molar concentrations (0.2 nM) are depicted in the autoradiographs of denaturing polyacrylamide gels shown in Figure 2.
Figure 2.
NER dual incisions of the 32P-internally labeled covalently closed circular plasmid (A), linearized plasmids (B), and 147 bp linear DNA duplexes (A) containing the same molar concentrations of DNA molecules bearing single (+)-cis-B[a]P-dG lesions (0.2 nM) as a function of incubation time in identical HeLa cell extracts (lanes labelled 0, 2, 5, 10, or 30 min). Lanes M: oligonucleotide size markers.
In the case of circular plasmids, the characteristic ladder of NER dual incision products of the 32P-labeled oligonucleotide fragments ~ 24 – 32 nt in lengths becomes clearly detectable within 5 min after mixing (Panel A, lane 10); the shorter oligonucleotide fragments are due to the partial degradation of the dual incision products catalyzed by exonucleases. The NER yields are significantly smaller in the case of the linearized plasmid and the dual incision products appear after 10 min (Panel B, lane 4). An even lower yield of dual incision products is observed in the case of 147 bp DNA duplexes under the same conditions (Panel A, lane 5). In the case of the 147 bp linear DNA duplexes, analogous exonuclease products are not observed up to ~2 h of incubation time, in agreement with previous experiments in human cell extracts.21, 22
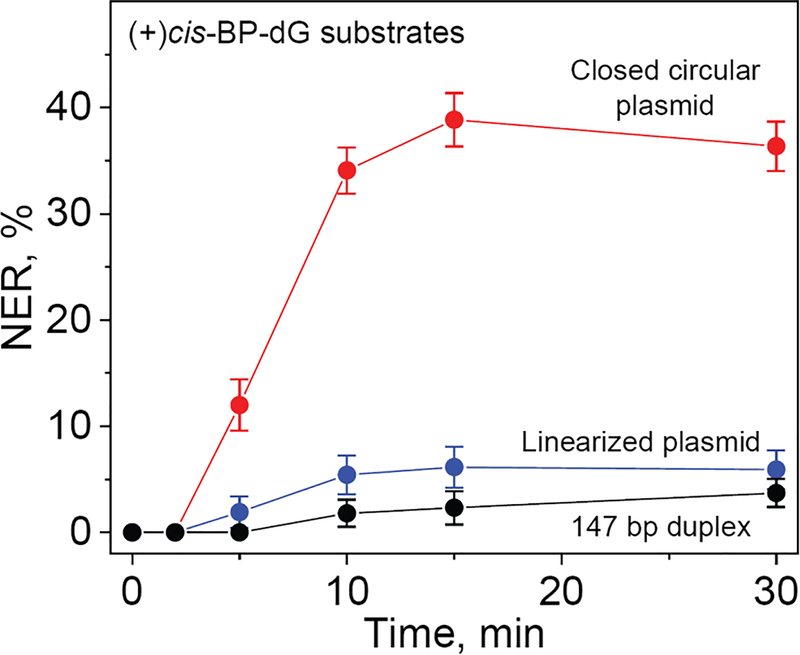
The NER dual incision kinetics observed in the case of the covalently closed circular plasmid, linearized plasmids, and 147 bp linear DNA duplexes are summarized in Figure 3. In the case of the plasmid substrates, the NER dual incision products appear within the first 5 min of incubation time and reach a level of 36–38% within 10 – 30 min. The linearized plasmid substrate is less reactive and attain a maximum level of about ~6% after a 30 min incubation time. The lowest NER activity was observed in the case of the 147 bp linear DNA substrates (~4%). These results demonstrate that the NER dual incision activity is the highest in the case covalently close circular plasmids.
Figure 3.
Kinetics of appearance of NER dual incision products after incubation of the covalently closed circular plasmids, linearized plasmids, and 147 bp linear DNA duplexes containing the same single (+)-cis-B[a]P-dG lesion. The NER yields were calculated by integration of the histograms derived from the denaturing gel autoradiographs at each particular incubation time (0, 2, 5, 10, 15 and 30 min). The results of three independent experiments and their standard deviations are shown.
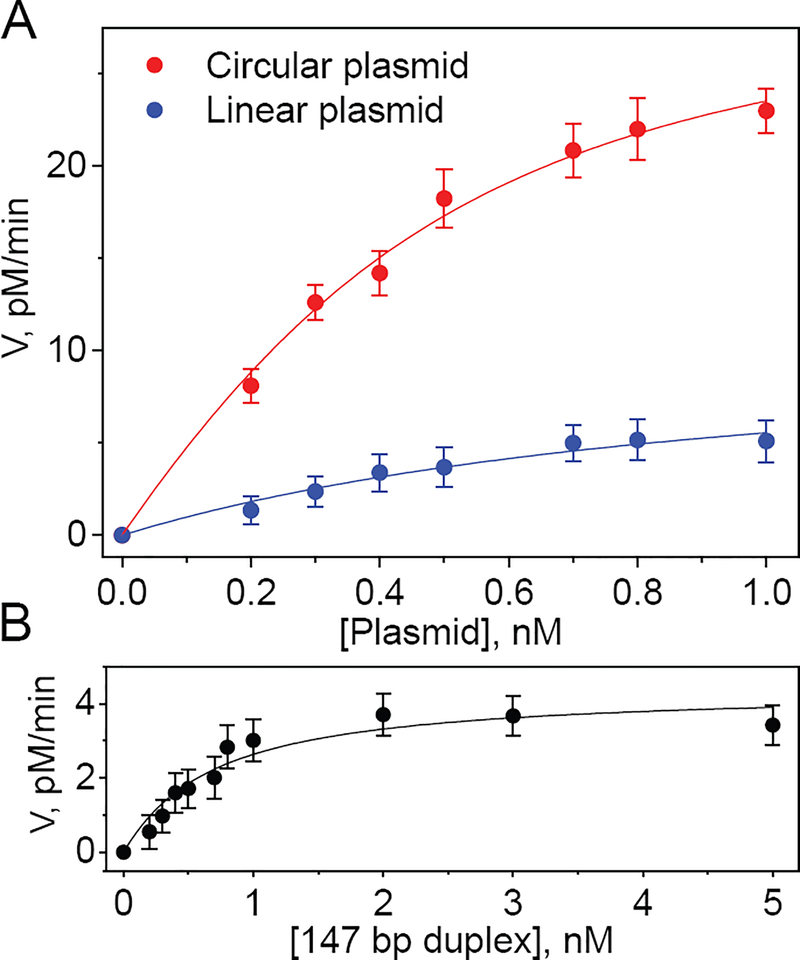
The low product yields of the linearized plasmid (linDNA) and 147-mers relative to the intact covalently closed plasmid (ccDNA) are not limited by low concentrations of NER factors since at least a six-fold higher concentration is available that leads to the efficient dual incision of the ccDNA (Figure 3). To obtain more insights into the factors that limit the yield of excision products, we examined the impact of varying DNA substrate concentrations on the rates of accumulation of NER dual incision products (Figure 4).
Figure 4.
Effect of concentration of covalently closed circular and linearized plasmids (A), and 147 bp linear DNA duplexes (B) containing single (+)-cis-B[a]P-dG lesions on the steady-state rate of appearance of NER dual incision products (V). The values of V were estimated using the initial data points beyond the apparent induction period (2 and 5 min, for the linearized plasmid and the 147 bp duplex, respectively, Figure 3). The solid lines represent the best fits of the following equation to the experimental data points with the standard errors representing 95% confidence intervals (the fitting parameters are summarized in Table 1) based on the equation that is valid over a wide range of substrate concentrations that represents an exact solution based on Scheme 1.
In all three cases the yields increase as a function of DNA concentration and tends to level off at the higher substrate concentrations according to the following Scheme 1:
Scheme 1.
Repair of damaged DNA substrates (D) by NER protein factor (FNER).
In this scheme, NER protein factor (FNER) binds to the damaged DNA substrates (D) to form the intermediate complex (FNER•D) with rate constant k1; the complexes either dissociate with rate constant k-1, or successfully undergo dual incisions to generate the NER dual incision products (PNER) with rate constant kP. These lesion-containing oligonucleotides are slowly released (~3.3 h) from the double-stranded DNA in a complex with the ten-protein NER factor TFIIH.22 The slow release of TFIIH on the time scale of our incubation in cell extracts (≤ 30 min) suggests that our NER kinetic experiments occur under single-turnover conditions. The steady-state rate of the product formation as a function of the substrate concentration can be expressed by the following equation:
| (1) |
where Kdiss = k−1/k1 is equal to the equilibrium dissociation constant of the FNER•D complex, because recycling of FNER under single-turnover conditions is negligible.23 At low substrate concentration [D] < Kdiss the value of V = kP[FNER][D]/Kdiss linearly increases with substrate concentration, whereas at high [D] the reaction becomes independent of [D] and asymptotically approaches its maximum rate, Vmax = kP[FNER] in good agreement with the experimental data (Figure 4). Applying such fitting routines yields excellent fits as shown by the solid lines in Figure 4, and the estimated Kdiss and Vmax values are summarized in Table 1.
Table 1.
The dissociation constants (Kdiss) and maximum rates (Vmax) for covalently closed DNA plasmid, linearized plasmid, and 147-mer linear duplex.
| Plasmid, ccDNA | Linearized Plasmid, linDNA | Linear DNA 147-mer duplex | |
|---|---|---|---|
| Kdiss (nM)a | 0.23±0.03 | 0.87±0.2 | 0.62±0.2 |
| Vmax (pM/min) | 30±3 | 11±2 | 4.4±0.8 |
Standard errors representing 95% confidence intervals.
The dissociation constant Kdiss = 0.23±0.03 nM in the case of ccDNA, is ~ 3 – 4 times smaller than in the case of linearized DNA and the linear 147-mer DNA duplexes. The extent of curvature within a region of the size of ~ 30–35 bp is unlikely to have a significant effect on the binding of a DNA damage-sensing protein of the of XPC-RAD23B (DNA footprint ~ 30–35 base pairs9). The values of the association constant k1 are thus likely to be similar in ccDNA and linDNA. Finally, the number of base pairs in ccDNA and linDNA are the same. Based on these considerations, we advance the hypothesis that the smaller Kdiss of the factor FNER is due to a lower dissociation constant k-1 (Scheme 1). The surprising implication of these findings is that the existence of the double-stranded termini, or ends in linDNA play a significant role in determining the magnitude of the NER dual incision yield.
The successful in vitro dual incisions require a minimum of NER factors that include, XPC, TFIIH, XPA, RPA, and the endonucleases XPF-ERCC1 and XPG (encoded by ERCC5).4 The general consensus is that in the case of Global Genomic NER, the multistep excision repair mechanism is initiated by the DNA damage sensor XPC, followed by the recruitment of the ten-protein factor TFIIH to the site of DNA damage, the unwinding of a 24 – 32 long oligonucleotide sequence by a process that involves the combined actions of XPB and XPD that is mediated by XPA and RPA, followed by the dual incisions catalyzed by the endonucleases XPF and XPG. The NER factor XPA plays a critical role in initiating the TFIIH-associated unwinding mechanism.
Our experiments do not provide any direct insights into the nature of NER factor FNER in the proposed mechanism (Scheme 1). The DNA damage sensing factor XPC is the critical NER-initiating factor that binds to unmodified DNA duplexes with a Kdiss of 1.1±0.2 nM, and in the case of a (+)-cis-B[a]P-dG lesion embedded in the middle of the same 50-mer duplexes, Kdiss = 0.41±0.10 nM (publication in preparation). The NER factor XPA binds to modified and unmodified DNA, but with more than ~ 100–1100 fold smaller affinities24, 25 than XPC. It has been suggested that the binding of XPA to the DNA-XPC-TFIIH complex is the rate-determining step in the NER dual incision multi-step mechanism.26 However, our Kdiss value of 0.23 nM (Table 1) is orders of magnitude smaller than the dissociation constants of XPA, while the Kdiss value extracted from the analysis of the data in Figure 4, suggests a value of 0.23 nM which is close to the XPC Kdiss value of 0.41 nM measured independently. While the importance of the other NER factors, besides XPA, cannot be rigorously excluded, it is likely that FNER ≈ XPC because it is the NER initiating factor that binds rapidly to unmodified and modified DNA,27 is known to scan DNA lesions by a long-range26 constrained one-dimensional diffusion mechanism,25, 28 and because it is consistent with the results summarized in Figure 4 and Table 1.
Impact of DNA termini and non-specific DNA binding of NER factors on dual incision yields
The dissociation constants of XPC to unmodified and cis-B[a]P-dG lesions are within a factor of less than two from one another.29 We therefore investigated whether the ratio between specific and non-specific binding decreases as a function of length of the DNA molecules containing a single B[a]P-dG lesion.
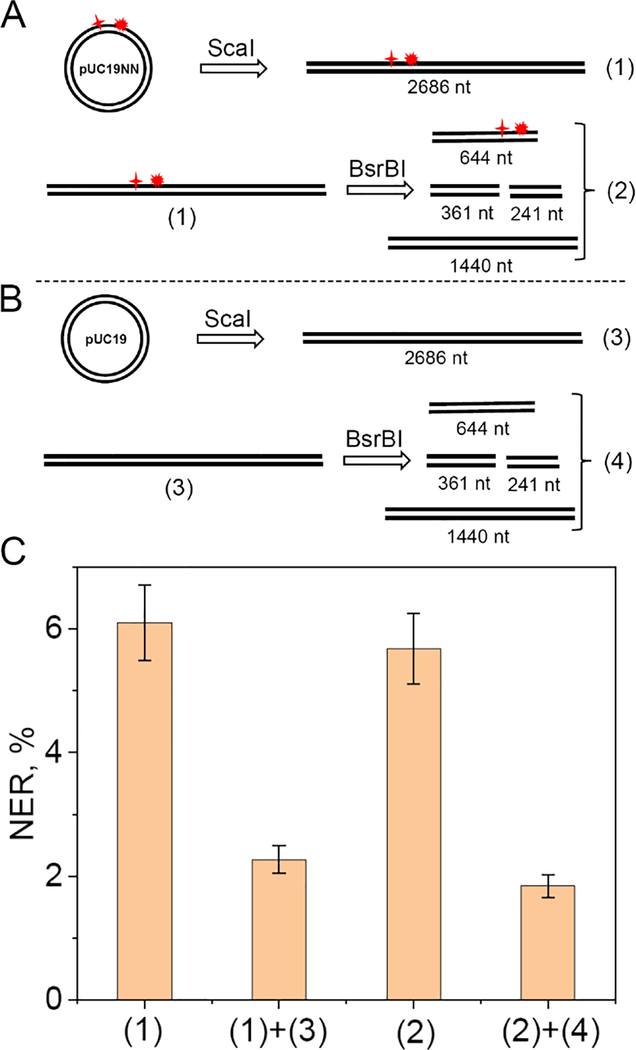
To test this hypothesis, we explored the effects of non-specific linear DNA molecule competitors on the yields of NER dual incision products. The pUC19NN plasmid was digested by the BsrBI restriction enzyme that has three specific cleavage sites, thus generating a mixture of four linear DNA fragments with blunt ends. The results of these experiments are summarized in Figure 5.
Figure 5.
Effects of non-specific linear DNA competitors on the yields of NER dual incision products with linearized plasmid substrates containing single (+)-cis-B[a]P-dG lesions. (A) Digestion of the circular 32P-labeled plasmid by ScaI to form the linearized plasmid substrate (1); digestion of the linearized substrate (1) with BsrBI to form a mixture of linear fragments (2). (B) Digestion of the non-radioactive pUC19 plasmid by ScaI to form the linearized plasmid (3); digestion of the linearized plasmid (3) by BsrBI to form a mixture of linear DNA fragments (4). (C) NER dual incision product yield after incubation of the linear DNA substrate (1) or (2), in the presence of non-specific competitor DNA fragments (3), or (4), derived from the digestion of 0.2 nM DNA substrates (1) or (3) as shown in panels A and B. Incubation time: 30 min in each case.
Unmodified competitor DNA sequences diminish NER yields
(1)+(3)
In the presence of equal concentrations (0.2 nM) of the unmodified DNA blunt-end sequence (3), derived from the treatment of the ccDNA plasmid with ScaI, the NER yield of (1) is reduced by a factor of 2.7 (Figure 5C). This experiment suggests that the availability of NER factors to DNA sequence (1) is reduced by the presence of an unmodified DNA molecule of equal length.
Comparing (1) and (2)
Fully digesting (1) with BsrBI to the set of fragments (2) shown in Figure 5A, each fragment present with concentrations of 0.2 nM, does not significantly affect the NER yields as shown by (2) in the bar graph (Figure 5C). This experiment suggests that the assembly of the NER complex in the case of the shorter 644 nt fragment with an efficiency similar to that in the longer 2686 fragment. Furthermore, increasing the number of DNA ends in mixture (2) by factor of 4, in comparison with the parent linearized plasmid (1), does not affect the yields of NER dual incision efficiencies.
(2)+(4)
Adding the mixture of unmodified fragments (4) derived from the digestion of the unmodified linear plasmid (1) to mixture (2), diminishes the NER yield by an additional factor of ~3.
Taken together, these results indicate that the reduction of NER yields are due more to the overall total concentrations of base pairs, rather than the number of molecules and thus the number of DNA duplex termini.
SUMMARY AND PERSPECTIVES
The remarkably high NER dual incision activities in HeLa cell extracts in the case of covalently closed plasmid DNA relative to linearized DNA by a factor of ~6 described here, is correlated with the reduction of NER repair synthesis activity by up to 80% in the case of linearized plasmid when compared with supercoil plasmid DNA.30 In the latter experiments, plasmids contained multiple lesions such as cyclobutane pyrimidine dimers, platinum, acetylaminofluorene and 8-methoxypsoralen adducts. Other experiments demonstrated that UV photoproducts in plasmid DNA microinjected in frog oocytes were repaired 50 times more rapidly in circular molecules than in linear DNA.31 However, neither contribution proposed a mechanistic explanation for these observations.
In this contribution, we propose that the striking elevated efficiency of removal a single bulky DNA lesion embedded in a covalently closed NER plasmid with a contour length of 2686 base pairs. This effect is attributed to the absence of termini that diminish the dissociation rate of one or more critical NER factors. This conclusion is supported by the ~ six-fold lower NER incision product yields in the linearized form of the same plasmid molecules. The nature of the critical NER factor has not been rigorously identified in this work. However, the working hypothesis, based on several important observations, suggests that the DNA damage sensor XPC-RAD23B factor is the most likely NER factor that can account for these observations. This NER damage-sensing NER factor is known to initiate the multi-step NER mechanism by binding first to the DNA lesions and subsequently recruiting other NER factors to the XPC-RAD23B-DNA complex. The search for DNA lesions occurs by a long-range, constrained one-dimensional scanning,25, 28 and/or translocation/hopping search mechanisms32–34 that need to be further investigated.
Supplementary Material
Acknowledgments
Funding
This work was supported by the National Institute of Environmental Health Sciences grant R01 ES-027059 to V.S. and R21 ES-028546 to N. E. G.
ABBREVIATIONS
- BER
base excision repair
- NER
nucleotide excision repair
- B[a]P-dG
10R (+)-cis-anti-B[a]PDE-N2-dG adduct
- (+)-7R,8S-dihydrodiol, B[a]PDE, 9S
10R-epoxy-tetrahydrobenzo[a]pyrene enantiomer
- bp
base pair
- nt
nucleotide
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
Figure S1 shows Sanger sequencing of pUC19NN plasmid.
REFERENCES
- [1].Lomax ME, Folkes LK, and O’Neill P (2013) Biological consequences of radiation-induced DNA damage: relevance to radiotherapy, Clin. Oncol 25, 578–585. [DOI] [PubMed] [Google Scholar]
- [2].Geacintov NE, and Broyde S (2017) Repair-Resistant DNA Lesions, Chem. Res. Toxicol. 30, 1517–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Torgovnick A, and Schumacher B (2015) DNA repair mechanisms in cancer development and therapy, Front. Genet 6, 157–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Marteijn JA, Lans H, Vermeulen W, and Hoeijmakers JH (2014) Understanding nucleotide excision repair and its roles in cancer and ageing, Nat. Rev. Mol. Cell. Biol. 15, 465–481. [DOI] [PubMed] [Google Scholar]
- [5].Huang JC, and Sancar A (1994) Determination of minimum substrate size for human excinuclease, J. Biol. Chem. 269, 19034–19040. [PubMed] [Google Scholar]
- [6].Hess MT, Schwitter U, Petretta M, Giese B, and Naegeli H (1997) Bipartite substrate discrimination by human nucleotide excision repair, Proc. Natl. Acad. Sci. U. S. A. 94, 6664–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mu D, and Sancar A (1997) DNA excision repair assays, Prog. Nucleic Acid Res. Mol. Biol. 56, 63–81. [DOI] [PubMed] [Google Scholar]
- [8].Mason TM, Smeaton MB, Cheung JC, Hanakahi LA, and Miller PS (2008) End modification of a linear DNA duplex enhances NER-mediated excision of an internal Pt(II)-lesion, Bioconjug. Chem 19, 1064–1070. [DOI] [PubMed] [Google Scholar]
- [9].Sugasawa K, Shimizu Y, Iwai S, and Hanaoka F (2002) A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex, DNA Repair (Amst) 1, 95–107. [DOI] [PubMed] [Google Scholar]
- [10].Wang H, and Hays JB (2001) Simple and rapid preparation of gapped plasmid DNA for incorporation of oligomers containing specific DNA lesions, Mol. Biotechnol. 19, 133–140. [DOI] [PubMed] [Google Scholar]
- [11].Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, and Egly JM (2007) The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions, EMBO J. 26, 2923–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hess MT, Gunz D, Luneva N, Geacintov NE, and Naegeli H (1997) Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes, Mol. Cell. Biol. 17, 7069–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shivji MK, Moggs JG, Kuraoka I, and Wood RD (2006) Assaying for the dual incisions of nucleotide excision repair using DNA with a lesion at a specific site, Method. Mol. Biol. 314, 435–456. [DOI] [PubMed] [Google Scholar]
- [14].Lowary PT, and Widom J (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning, J. Mol. Biol. 276, 19–42. [DOI] [PubMed] [Google Scholar]
- [15].Shafirovich V, Kolbanovskiy M, Kropachev K, Liu Z, Cai Y, Terzidis MA, Masi A, Chatgilialoglu C, Amin S, Dadali A, Broyde S, and Geacintov NE (2019) Nucleotide excision repair and impact of site-specific 5′,8-cyclopurine and bulky DNA lesions on the physical properties of nucleosomes, Biochemistry 58, 561–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Smeaton MB, Miller PS, Ketner G, and Hanakahi LA (2007) Small-scale extracts for the study of nucleotide excision repair and non-homologous end joining, Nucleic Acids Res. 35, e152–e152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kang TH, Reardon JT, Kemp M, and Sancar A (2009) Circadian oscillation of nucleotide excision repair in mammalian brain, Proc. Natl. Acad. Sci. U. S. A. 106, 2864–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Huang JC, Hsu DS, Kazantsev A, and Sancar A (1994) Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches, and bulky adducts, Proc. Natl. Acad. Sci. U. S. A. 91, 12213–12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gillet LC, and Scharer OD (2006) Molecular mechanisms of mammalian global genome nucleotide excision repair, Chem. Rev. 106, 253–276. [DOI] [PubMed] [Google Scholar]
- [20].Conney AH (1982) Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture, Cancer Res. 42, 4875–4917. [PubMed] [Google Scholar]
- [21].Svoboda DL, Taylor JS, Hearst JE, and Sancar A (1993) DNA repair by eukaryotic nucleotide excision nuclease. Removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by Xenopus laevis oocytes, J. Biol. Chem. 268, 1931–1936. [PubMed] [Google Scholar]
- [22].Kemp MG, Reardon JT, Lindsey-Boltz LA, and Sancar A (2012) Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair, J. Biol. Chem. 287, 22889–22899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Michaelis L, and Menten ML (1913) Die Kinetik der Invertinwirkung, Biochem. Z. 49, 333–369. [Google Scholar]
- [24].Hey T, Lipps G, and Krauss G (2001) Binding of XPA and RPA to damaged DNA investigated by fluorescence anisotropy, Biochemistry 40, 2901–2910. [DOI] [PubMed] [Google Scholar]
- [25].Beckwitt EC, Jang S, Carnaval Detweiler I, Kuper J, Sauer F, Simon N, Bretzler J, Watkins SC, Carell T, Kisker C, and Van Houten B (2020) Single molecule analysis reveals monomeric XPA bends DNA and undergoes episodic linear diffusion during damage search, Nat. Commun 11, 1356–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sugasawa K, Akagi J, Nishi R, Iwai S, and Hanaoka F (2009) Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning, Mol Cell 36, 642–653. [DOI] [PubMed] [Google Scholar]
- [27].Trego KS, and Turchi JJ (2006) Pre-steady-state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination, Biochemistry 45, 1961–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cheon NY, Kim HS, Yeo JE, Scharer OD, and Lee JY (2019) Single-molecule visualization reveals the damage search mechanism for the human NER protein XPC-RAD23B, Nucleic Acids Res. 47, 8337–8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lee YC, Cai Y, Mu H, Broyde S, Amin S, Chen X, Min JH, and Geacintov NE (2014) The relationships between XPC binding to conformationally diverse DNA adducts and their excision by the human NER system: Is there a correlation?, DNA Repair (Amst) 19, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Calsou P, Frit P, and Salles B (1996) Double strand breaks in DNA inhibit nucleotide excision repair in vitro, J. Biol. Chem. 271, 27601–27607. [DOI] [PubMed] [Google Scholar]
- [31].Legerski RJ, Penkala JE, Peterson CA, and Wright DA (1987) Repair of UV-induced lesions in Xenopus laevis oocytes, Mol. Cell. Biol. 7, 4317–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Halford SE, and Marko JF (2004) How do site-specific DNA-binding proteins find their targets?, Nucleic Acids Res 32, 3040–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mirny L, Slutsky M, Wunderlich Z, Tafvizi A, Leith J, and Kosmrlj A (2009) How a Protein Searches for Its Site on DNA: The Mechanism of Facilitated Diffusion, J. Phys. A: Math. Theor. 42, 434013–434013. [Google Scholar]
- [34].Esadze A, and Stivers JT (2018) Facilitated Diffusion Mechanisms in DNA Base Excision Repair and Transcriptional Activation, Chem. Rev. 118, 11298–11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.