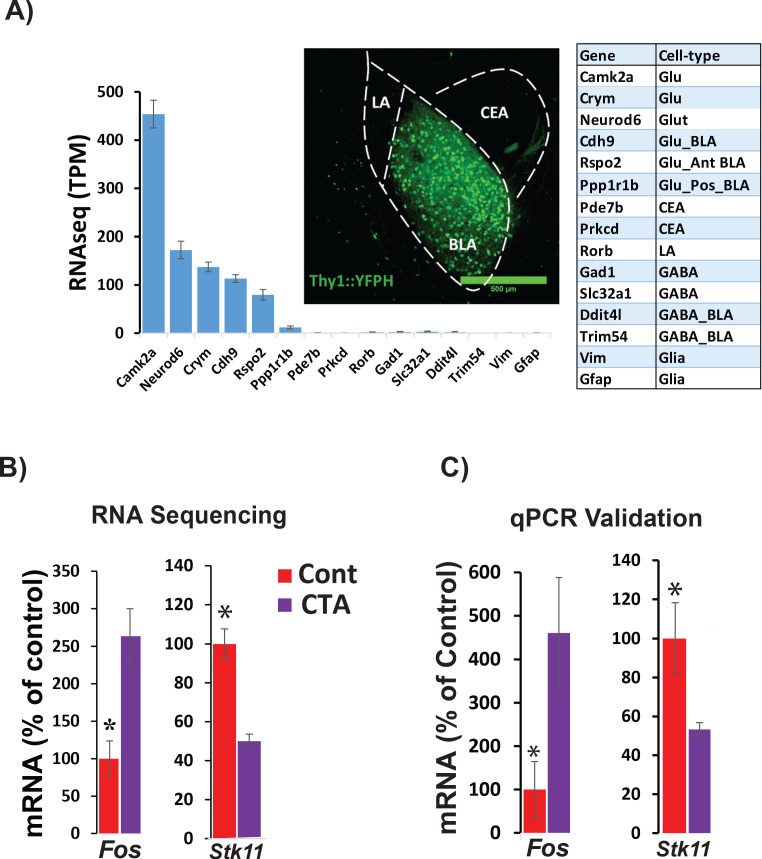
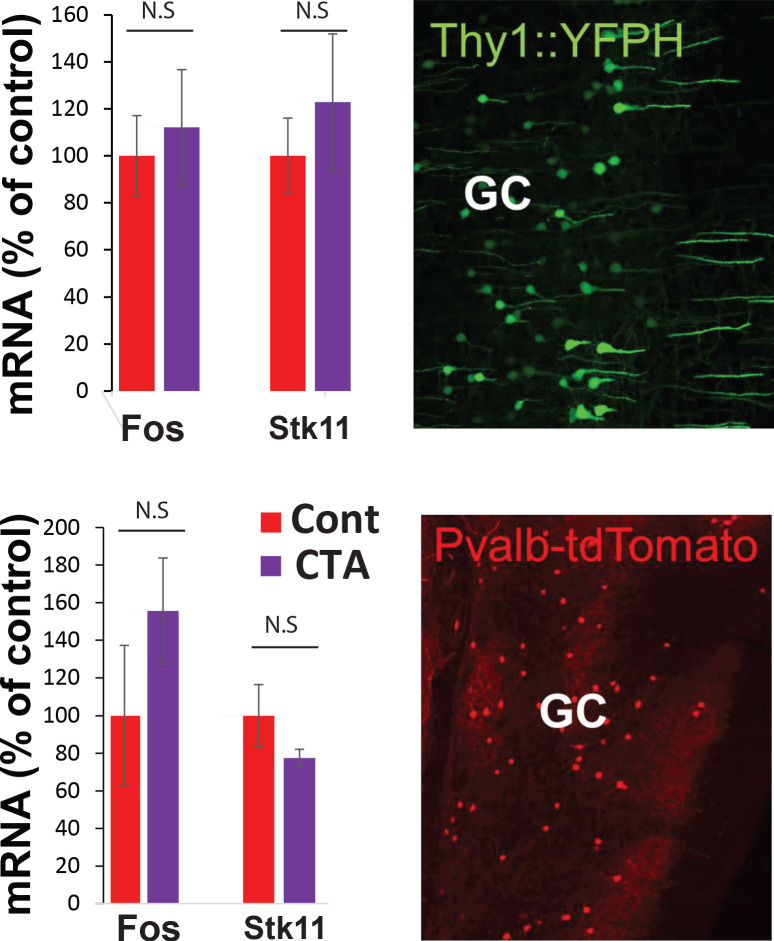
Figure 2. RNA sequencing from BLApn 4 hr following CTA training.
(A) BLApn were isolated from YFP-H mice following CTA or taste control (N = 4/group). Neurons (150-200) were manually sorted form coronal slices (LA-lateral amygdala; CEA-central amygdala). Abundant transcripts (histogram, averaged across both groups) are enriched for those expected in the population and depleted for those expressed in other nearby populations (Table 1) including GABAergic interneurons, glia, or neurons in LA or CEA; Sugino et al., 2006; Kim et al., 2016; Allen brain atlas). Glu- GABA-, glutamatergic, GABAergic neurons; AntBLA, PostBLA- Anterior and posterior portions of the BLA. TPM- transcript per million. (B) Among genes meeting robust criteria for differential expression (see Table 1) Stk11 and Fos were selected for further analysis, including qPCR confirmation (C) in separate experiments (N = 4/group; *p<0.05). See also Figure 2—source data 1 and Figure 2—figure supplements 1–4 and Tables 1–3.
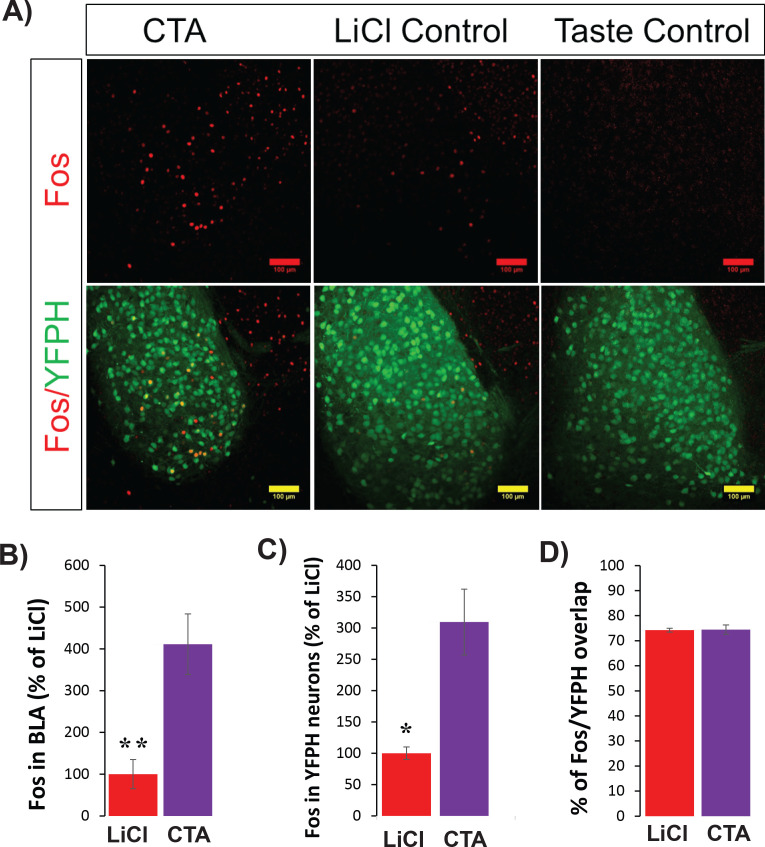
Figure 2—figure supplement 1. CTA increases C-FOS protein expression in BLApn including those in strain YFP-H.
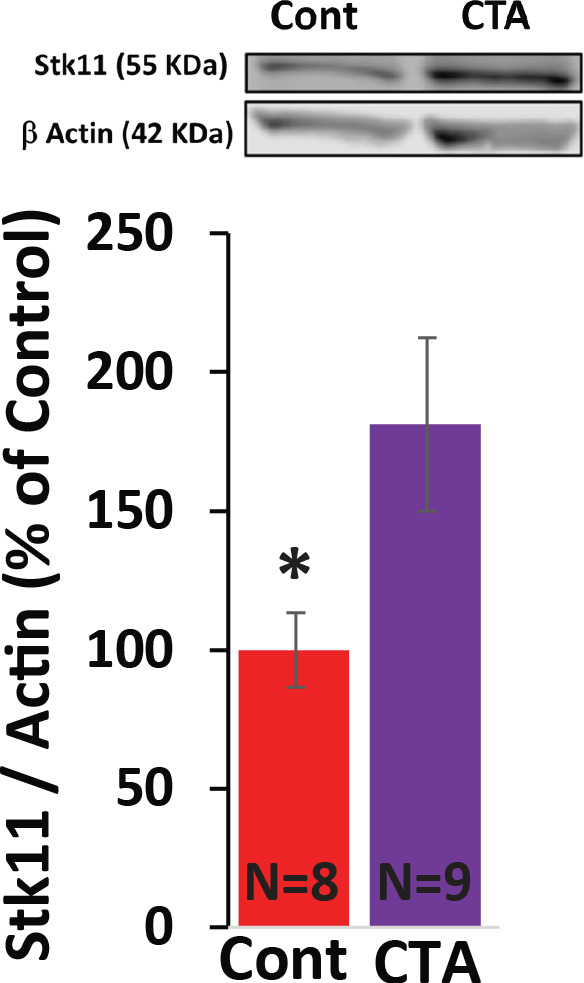
Figure 2—figure supplement 2. STK11 protein expression following CTA training.