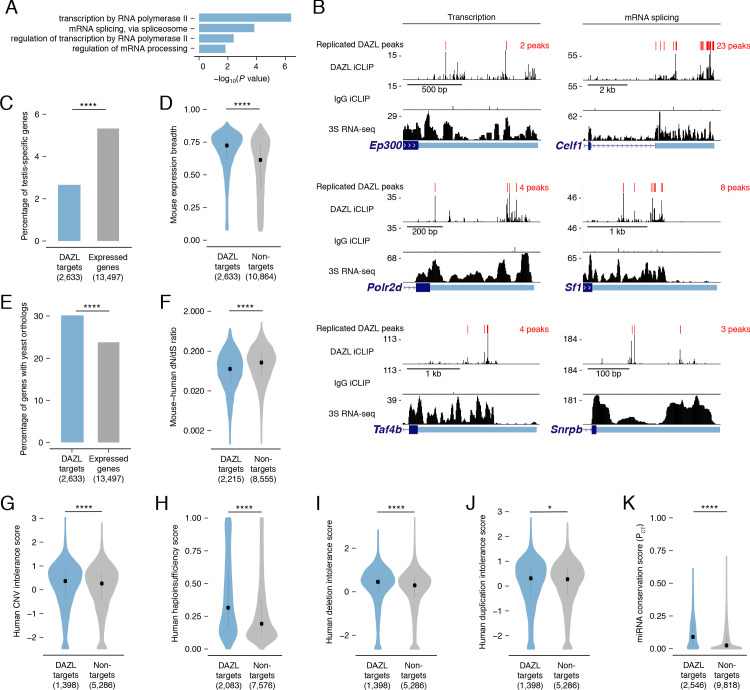
Figure 3. DAZL targets are enriched for broadly expressed, dosage-sensitive regulators of basic cellular processes.
(A) All statistically enriched GO biological processes, excluding parental nodes, for 2,633 DAZL-targeted genes compared with all genes expressed in undifferentiated spermatogonia (TPM ≥1) using the PANTHER GO Slim annotation set. P values are from binomial tests with Bonferroni correction. (B) DAZL iCLIP, IgG iCLIP, and 3S RNA-seq gene tracks showing exemplary DAZL-bound genes that are involved in transcription and RNA splicing. Gene tracks as described in Figure 2E. (C) DAZL targets are depleted of testis-specific factors compared with all genes expressed in undifferentiated spermatogonia (one-tailed hypergeometric test). (D) DAZL targets are more broadly expressed than nontargets within 12 adult mouse tissues (one-sided Mann-Whitney U test). (E) DAZL targets are more likely to have a yeast ortholog (one-tailed hypergeometric test). (F) DAZL targets have a reduced ratio of nonsynonymous substitutions per nonsynonymous site to synonymous substitutions per synonymous site (dN/dS) in alignments with human orthologs (one-sided Mann-Whitney U test). (G) Human orthologs of DAZL’s targets exhibit increased intolerance for copy number variation (CNV) (one-sided Mann-Whitney U test). (H) The human orthologs of DAZL’s targets exhibit increased probability of haploinsufficiency (one-sided Mann-Whitney U test). (I) The human orthologs of DAZL’s targets exhibit increased intolerance for deletions (one-sided Mann-Whitney U test). (J) The human orthologs of DAZL’s targets exhibit increased intolerance for duplications (one-sided Mann-Whitney U test). (K) DAZL targets exhibit higher miRNA conservation scores (PCT) (one-sided Mann-Whitney U test). Violin plots display medians with interquartile ranges; number of genes (n) in each group designated in parentheses in labels along x-axis. *, p<0.05; ****, p<0.0001.