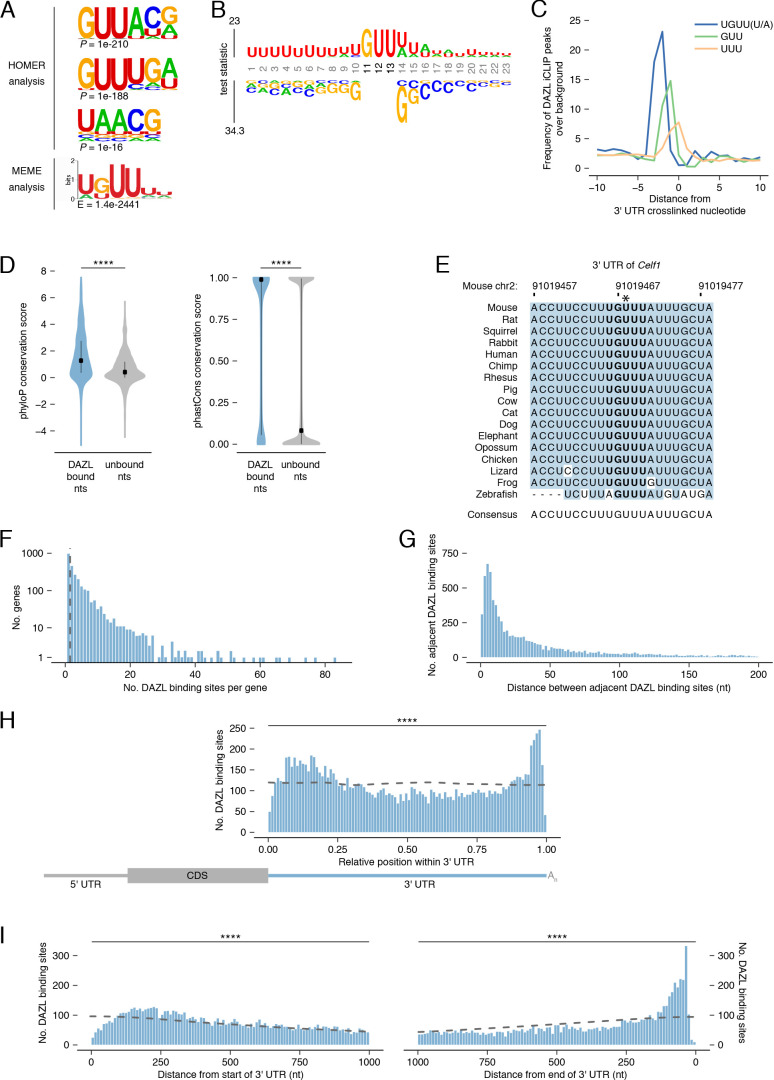
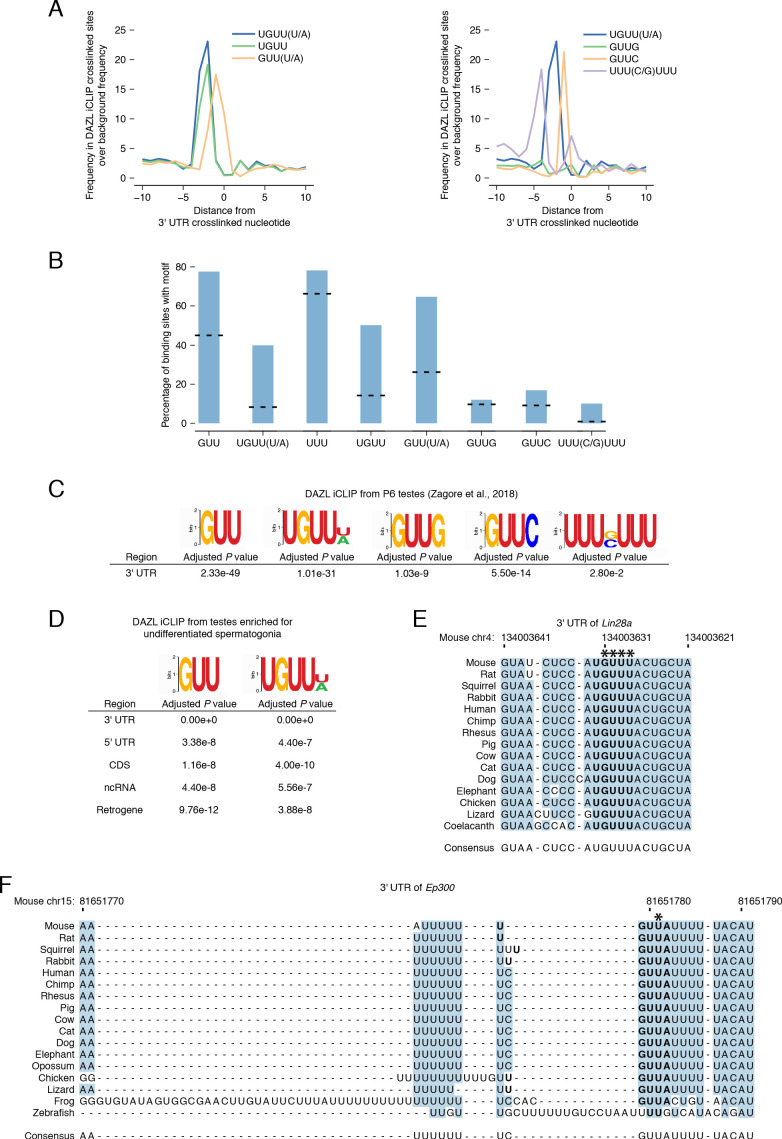
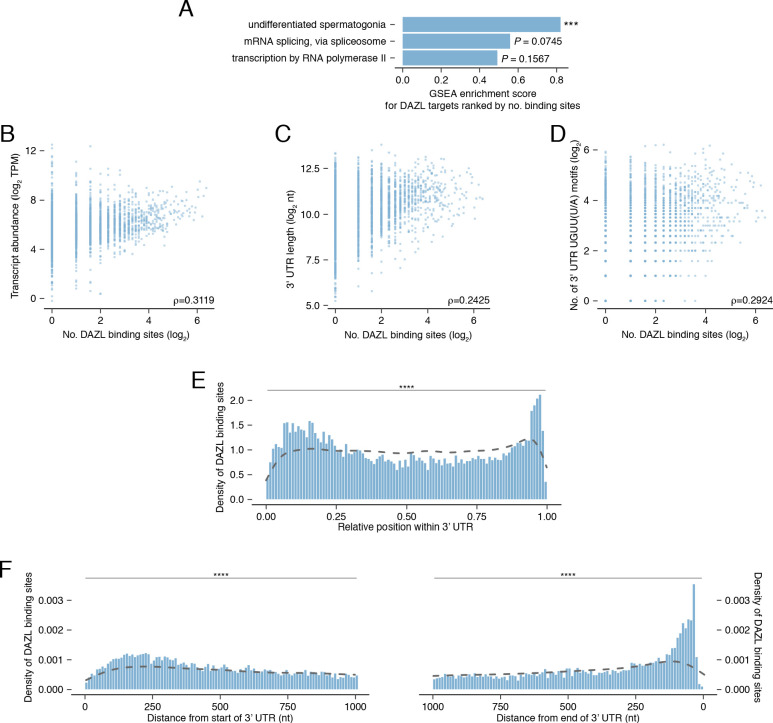
Figure 4. DAZL binds a UGUU(U/A) motif within 3' UTRs.
(A) De novo motif discovery from replicated DAZL peaks in 3' UTR exons. Motif analyses were carried out with HOMER and MEME tools using crosslinked peaks ± 10 nucleotides, with all expressed 3' UTRs (TPM ≥1) as background. The top three ranked motifs identified via HOMER are shown. One statistically significant motif was identified via MEME. (B) GUU-centered motif analysis of replicated peaks in 3' UTRs via kpLogo. For each crosslinked peak ±10 nucleotides, the closest GUU was identified, and all sequences were aligned along the GUU. Background was a subset of unbound GUUs randomly selected sequences from the full-length 3' UTRs that contain DAZL peaks. As P values are extremely small (<1×10−308), residues are scaled by test statistics. (C) Position of all UGUU(U/A), GUU, and UUU motifs relative to crosslinked nucleotides from replicated peaks in 3' UTRs. 0 represents the crosslinked nucleotide. Enrichment was identified relative to randomly selected sequences from the full-length 3' UTRs that contain DAZL peaks. (D) Conservation of DAZL binding sites across vertebrates based on phyloP and phastCons scores. DAZL-bound nucleotides identified via iCLIP were compared with unbound nucleotides from the same 3' UTRs (two-sided Mann-Whitney U test). (E) DAZL’s 3' UTR binding site in Celf1 is conserved among vertebrates. Blue shading highlights nucleotides that reflect the consensus. Bold designates DAZL’s UGUU(U/A) motif. Asterisk marks crosslinked nucleotide in DAZL iCLIP data. Sequence shown is absent from coelacanth. (F) Frequency of DAZL binding sites per DAZL-bound transcript. The majority of DAZL targets have more than one DAZL binding site (those targets to the right of the vertical dashed line). (G) Distance between adjacent DAZL binding sites in 1,649 DAZL-bound transcripts with more than one DAZL binding site. (H) Relative position of DAZL binding sites along the 3' UTR. The start and end of the 3' UTR were designated as 0 and 1, respectively. DAZL binding sites are enriched at the end and, to a lesser extent, at the start, relative to randomly selected sites in the same 3' UTRs (dashed line) (two-sided Kolmogorov-Smirnov test). (I) Absolute position of DAZL binding sites along the 3' UTR. DAZL binding sites exhibit a sharp accumulation 20–100 nucleotides from the end of the 3' UTR and a broader accumulation 100–240 nucleotides from the start of the 3' UTR relative to randomly selected positions within the same 3' UTRs (dashed line) (two-sided Kolmogorov-Smirnov tests). ****, p<0.0001.