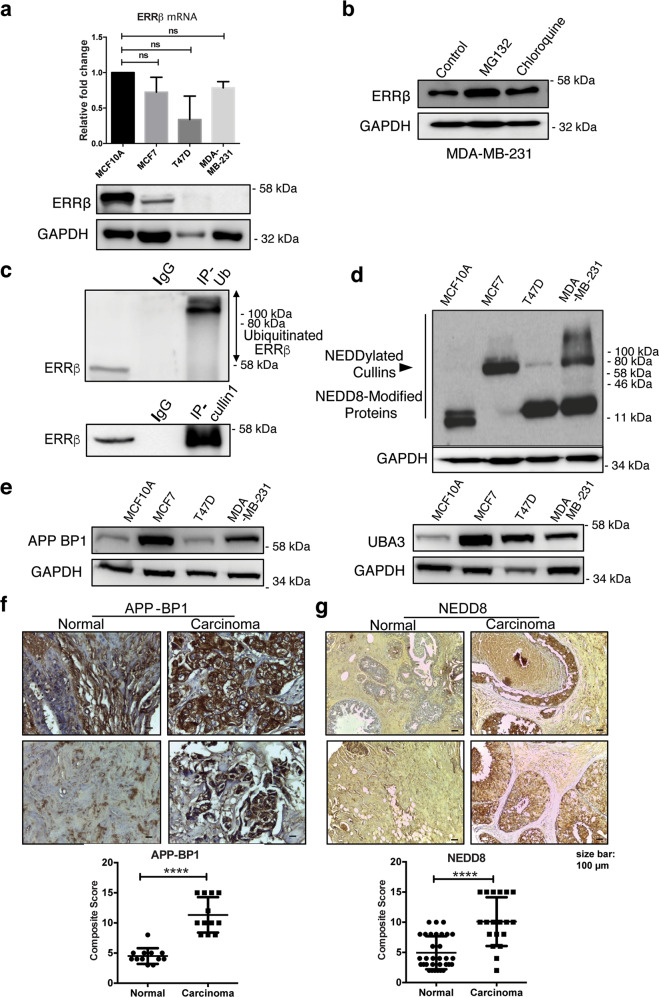
Fig. 1. ERRβ protein expression is downregulated by NEDD8 activity in breast cancer.
a Relative ERRβ mRNA expression level of MCF10A, MCF7, T47D and MDA-MB-231 cells were analysed using qRT-PCR. Three technical repeats (N = 3) were performed and the data represent the means ± SD. (two-tailed t test; Significant: ns no significant difference) (upper panel). Representative western blot analysis of the ERRβ in MCF10A, MCF7, T47D and MDA-MB-231 cells. GAPDH was used as a loading control (lower panel). b MDA-MB-231 cells were treated with 1 µM MG132 and Chloroquine independently for 12 h and the ERRβ protein expression was analysed by western blotting. Representative Western blot is shown. GAPDH was used as a loading control. (N = 3). c Co-immunoprecipitation was performed with antibodies against Ubiquitin (Ub) and Cullin1 and analysed with ERRβ antibodies using western blotting. Anti-IgG antibody was used as a negative control. d Western blot analysis of the NEDD8 in in MCF7, MCF10A, T47D and MDA-MB-231 cells. GAPDH was used as a loading control. (N = 3). e APP-BP1 and UBA3 expression was analysed by western blotting in MCF10A, MCF7, T47D and MDA-MB-231 cells. GAPDH was used as a loading control. (N = 3). f, g Immunohistochemical (IHC) staining of normal (left) and carcinoma (right) breast tissue samples using the APP-BP1 and NEDD8 antibody, respectively. Graphical representation (below) was the IHC composite score of each tissue microarray sample. A composite score (<3 = low; 3–5 = moderately; ≥6 = highly categorised) was calculated for each sample using intensity score and percentage of APP-BP1 and and NEDD8 stained-positive cells. (2-sample t test; significant: ****p < 0.001, very significant). Bar = 100 μm.