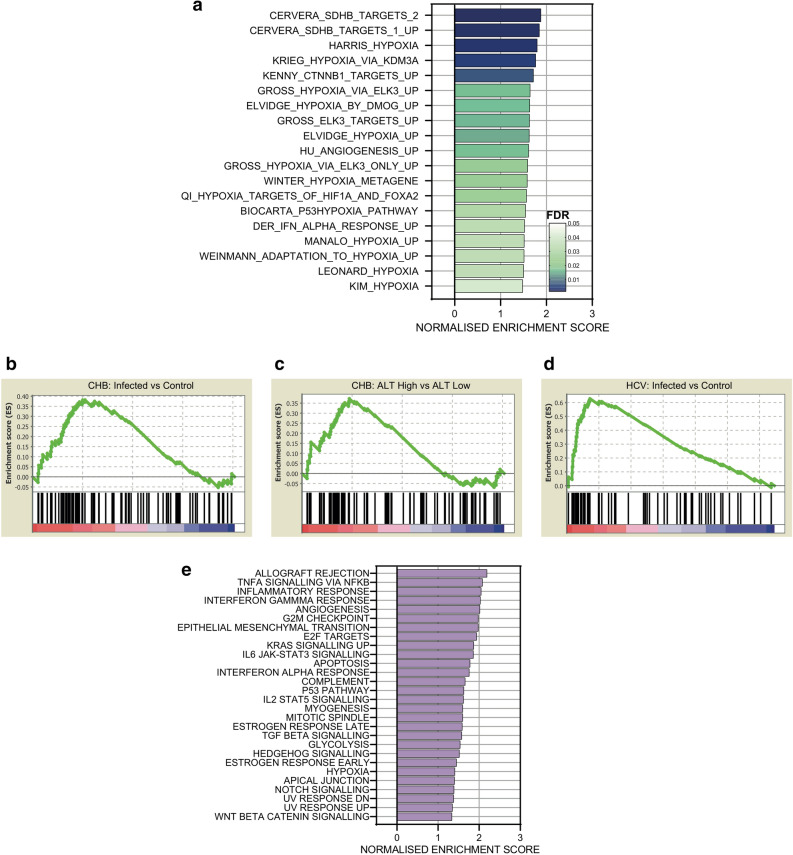
Figure 1.
Increased hypoxia gene expression in CHB. Hypoxia upregulated gene signatures from Molecular Signatures Database were assessed in the CHB cohort, 19 significantly upregulated gene signatures identified (FDR < 0.05) and ranked by Normalized Enrichment Score (NES) (a). GSEA shows a significant enrichment of HepG2 defined hypoxic genes in CHB cohort (FDR = 0.077). The gene set was based on Fold Change > 2, and FDR < 0.05; 80 genes satisfied these criteria and are listed in Supplementary Table 2 (b). CHB cohort was grouped by peripheral ALT activity, with subjects > 40 IU/L (n = 57) or < 40 IU/L (n = 25). GSEA shows a significant enrichment of HepG2 defined hypoxic genes in patients with elevated ALT (FDR = 0.110) (c). HepG2 hypoxic gene set was enriched (FDR = 0.006) in HCV infected patients with cirrhosis (n = 41) compared to normal liver controls (n = 19) (d). MSigDB hallmark gene sets identified the most upregulated pathways in the CHB cohort: 28 gene sets were significantly enriched (FDR < 0.05) and are ranked by NES (e). All GSEA was performed using GSEA_4.0.371.