Abstract
Adaptive Immune responses generated by SARS-CoV-2 virus in convalescent patients according to disease severity remain poorly characterized. To this end, we designed a prospective study (NCT04365322) that included 60 COVID-19 convalescent patients (1-month post infection) in two cohorts respectively entitled mild illness and severe pneumonia. The monitoring of peripheral immune responses was performed using IFNᵧ ELISpot assay. The serology index of each patient was investigated at the same time. Patients with severe pneumonia were older and had more comorbidities than patients with mild illness. T-cell responses in term of frequency and intensity were clearly distinct between mild illness and severe pneumonia patients. Furthermore, our results demonstrated that recent history of COVID-19 did not hamper viral memory T-cell pool against common viruses (Cytomegalovirus, Epstein-Barr-virus and Flu-virus). The presence of potent adaptive immunity even in patients who underwent severe pneumonia sustain the rationale for the development of protective therapeutics against SARS-CoV-2.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Adaptive immunity, T-cells, Interferonᵧ
To the editor:
SARS-CoV-2 virus induces symptoms of variable severity; some patients only have mild illness whereas other rapidly become critically ill progressing to an acute respiratory distress syndrome (ARDS). This critical state of the disease supports the immediate relevance for the development of protective therapeutics against SARS-CoV-2 but requires fundamental knowledge concerning adaptive immune responses induced by the virus. Therefore we have read with interest the recent findings published by Bo and colleagues describing in the early stage of the disease a positive correlation between T-cells decrease and COVID-19 severity.1 However, Thijsen and colleagues demonstrated the presence of specific T-cell responses against S and N proteins few days post onset symptoms in patients with severe pneumonia hospitalized in intensive care unit (ICU).2 Although the existence of SARS-CoV-2 specific T-cells has been described,2 , 3 the frequency and the intensity of SARS-CoV-2 specific T-cell responses among mild illness and severe pneumonia convalescent COVID-19 patients remains to be investigated.
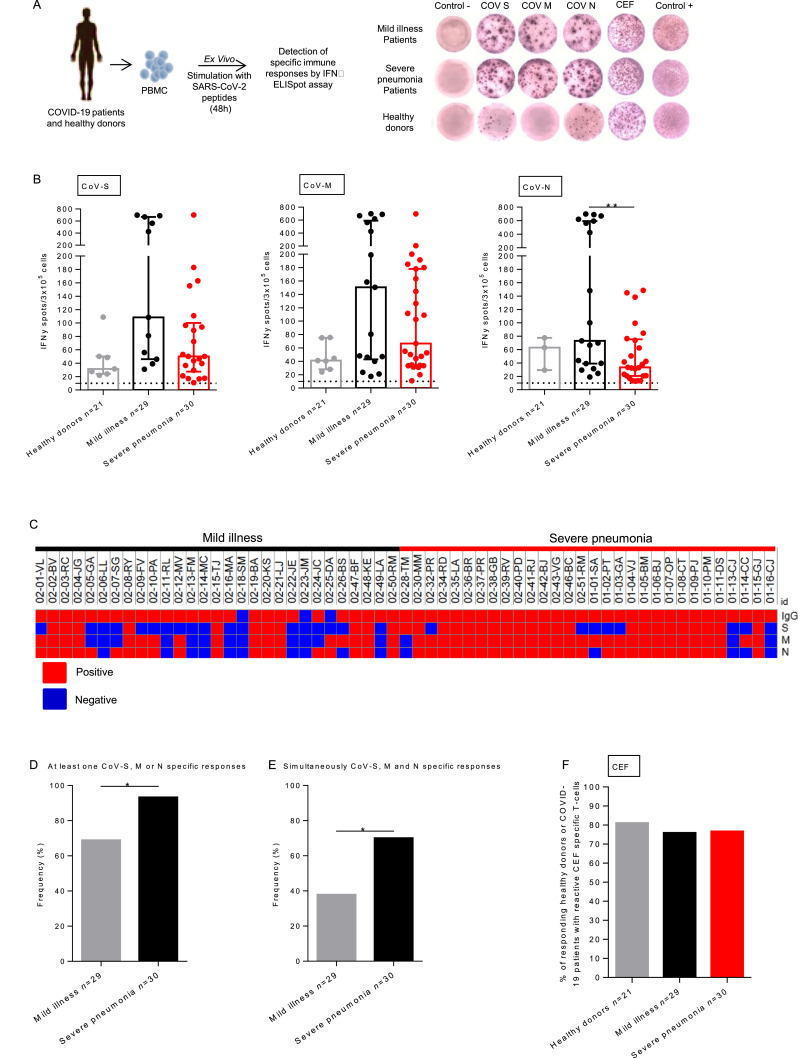
In this prospective study, 60 patients who had COVID-19 were enrolled in a two cohorts study that were entitled mild illness (n = 30) and severe pneumonia (n = 30) at least 21 days after the first symptoms of COVID-19 (38 days [21–53]; Table 1 ). The aim was to describe adaptive immune responses in COVID-19 convalescent patients according to their disease severity. All patients were considered as cured at the time of inclusion. Patients’ characteristics are detailed in Table 1. The mean age was 54.1 +/- 17.4, and 30 patients (50.0%) were male. Patients with severe pneumonia had more diabetes (20.0% vs 0.0%, P<0.001), hypertension (47.0% vs 6.7%, P<0.001), higher body mass index (24.1 vs 20.5, P<0.001), they were older (68.3 vs 39.8, P<0.001) and more frequently male (77.0% vs 23.0%) P<0,001) as previously described.4 Interestingly, patients with severe pneumonia had also more fever (86.7% vs 43.3%, P<0.001), dyspnea (86.7% vs 6.7% P<0.001) and less myalgia (16.7% vs 60.0%, P<0.001). Among patients who presented severe pneumonia, 23 (76.7%) were hospitalized in ICU for acute respiratory distress related to COVID-19. No patient died during the study period. The identification of SARS-CoV-2 specific T-cell responses was performed using ex vivo ELISpot assays that measure IFNᵧ produced by activated T-cells (Fig. 1 A). Median intensities for CoV-N T-cells responses were higher (P = 0.006) and the one of CoV-S (P = 0.056) and CoV-M (P = 0.29) tends to be increased in patients with mild illness compared to patients with severe pneumonia (73.0 IFNᵧ spot/3 × 105 cells [IQR: 39.0–595.0] vs 33.5 IFNᵧ spot/3 × 105 cells [IQR: 20.5–75.2]; 108.5 IFNᵧ spot/3 × 105 cells [IQR: 46.0–669.0] vs 50.0 IFNᵧ spot/3 × 105 cells [IQR: 27.4–100.1] and 150.5 IFNᵧ spot/3 × 105 cells [IQR: 43.0–591.8] vs 66.5 IFNᵧ spot/3 × 105 cells [IQR: 94.0–178.0] for CoV-N, CoV-S and CoV-M respectively; Fig. 1B). Additionally, T-cells’ specificity was analyzed based on IFNᵧ secretion using flow cytometry in thirteen patients from both cohorts (n = 6 mild illness patients and n = 7 severe pneumonia patients). Specific T-cell responses against S, M and N proteins were mediated by CD8 and CD4T-cells in both cohorts (data not shown). The presence of SARS-CoV-2 specific T-cells in healthy donors (33% for CoV-S and M and 14% for CoV-N) might be explained by the sequence homology between structural proteins from various coronavirus suggesting the existence of cross reactive memory T-cells.5 Indeed, high degrees of similarities between coronaviruses and SARS-CoV-2 concerning S, M and N structural proteins have been recently described.6 , 7
Table 1.
Characteristics of patients with COVID-19. Comparison between severe pneumonia and mild disease.
| Variables | Total (n = 60) | Mild disease (n = 30) | Severe pneumonia (n = 30) | Univariate P value |
|---|---|---|---|---|
| Male sex | 30 (50) | 7 (23.3) | 23 (76.7) | <0.001 |
| Mean age ±SD | 54.1 (±17.4) | 39.8 (±11.7) | 68.3 (±7.59) | <0.001 |
| Body mass index, mean ±SD | 22.3 (±4.23) | 20.5 (±3.72) | 24.1 (±3.99) | <0.001 |
| Comorbidities, n (%) | ||||
| Hypertension | 16 (26.7) | 2 (6.7) | 14 (46.7) | <0.001 |
| Chronic pulmonary disease | 8 (13.3) | 1 (3.3) | 7 (23.3) | 0.052 |
| Diabetes melitus | 6 (10) | 0 (0) | 6 (20) | 0.024 |
| Coronary heart disease | 1 (1.7) | 0 (0) | 1 (3.3) | 1 |
| Chronic renal insufficiency | 1 (1.7) | 0 (0) | 1 (3.3) | 1 |
| McCabe score | ||||
| Non fatal, n (%) | 39 (65) | 28 (93.3) | 11 (36.7) | <0.001 |
| Ultimately fatal, n (%) | 19 (31.7) | 2 (6.7) | 17 (56.7) | |
| Rapidly fatal, n (%) | 2 (3.3) | 0 (0) | 2 (6.7) | |
| Clinical characteristics, n (%) | ||||
| Fever | 39 (65) | 13 (43.3) | 26 (86.7) | <0.001 |
| Myalgia or arthralgias | 23 (38.3) | 18 (60) | 5 (16.7) | <0.001 |
| Fatigue | 44 (73.3) | 24 (80) | 20 (66.7) | 0.24 |
| Headache | 23 (38.3) | 15 (50) | 8 (26.7) | 0.063 |
| Diarrhea | 14 (23.3) | 3 (10) | 11 (36.7) | 0.015 |
| Dyspnea | 28 (46.7) | 2 (6.7) | 26 (86.7) | <0.001 |
| Sputum production | 12 (20) | 8 (26.7) | 4 (13.3) | 0.2 |
| Cough | 40 (66.7) | 18 (60) | 22 (73.3) | 0.27 |
| Hospitalisation, n (%) | 30 (50) | 0 | 30 (100) | – |
| Length of stay, dy mean ±SD | 26±15 | 26±15 | – | |
| Transfer in ICU, n (%) | 23 (38.3) | 0 | 23 (76.7) | – |
| Length of stay, dy mean ±SD | 19±12 | 19±12 | – | |
| ARDS, n (%) | 28 (46.7) | 28 (93.3) | ||
| Non specific therapy, n (%) | ||||
| Oxygenotherapy | 30 (50) | 0 | 30 (100) | – |
| Mechanical ventilation | 20 (33.3) | 20 (66.7) | – | |
| Neuromuscular blocking agent | 20 (33.3) | 20 (66.7) | – | |
| Prone positionning | 17 (28.3) | 17 (56.7) | – | |
| Inhaled nitric oxide | 2 (3) | 2 (6.7) | – | |
| Vasopressors | 11 (18.3) | 11 (36.7) | – | |
| Specific therapy, n (%) | 28 (46.7) | 0 | 28 (93.3) | – |
| Corticosteroids | 10 (16.7) | 10 (33.3) | – | |
| Interferon | 7 (11.7) | 7 (23.3) | – | |
| Lopinavir/ritonavir | 9 (15) | 9 (30) | – | |
| Hydroxychloroquine | 12 (20) | 12 (40) | – | |
| Remdesivir | 4 (6.7) | 4 (13.3) | – | |
| Laboratory findings, moy (±SD) | ||||
| Delay first symptom/blood sample, dy med [min-max] | 38 [21–53] | 39 [27–51] | 38 [21–45] | 0.019 |
| Lymphocyte count (G/L), mean (±SD) | 2.10 (±0.761) | 2.29 (±0.565) | 1.91 (±0.884) | <0.01 |
| Serology | ||||
| Positive IgG serology, n (%) | 57 (95) | 27 (90) | 30 (100) | 0.24 |
| Index, mean ±SD | 6.05 (±2.18) | 4.92 (±2.40) | 7.19 (±1.09) | <0.001 |
| T-cell responses*, n (%) | ||||
| CEF positivity | 45/59 (76.3) | 22/29 (75.9) | 23/30 (76.7) | 0.94 |
| COV M positivity | 44/59 (74.6) | 17/29 (58.6) | 27/30 (90) | <0.01 |
| COV N positivity | 43/59 (72.9) | 19/29 (65.5) | 25/30 (83.3) | 0.14 |
| COV S positivity | 33/59 (55.9) | 11/29 (37.9) | 22/30 (73.3) | <0.01 |
ELISpot was not performed for one patient, ARDS = Acute Respiratory distress syndrome.
Fig. 1.
SARS-CoV-2 specific T-cell responses were increased in mild illness compared to severe pneumonia COVID-19 patients. A. PBMC from 21 healthy donors and COVID-19 patients with mild illness (n = 29) or severe pneumonia (n = 30) were analyzed for SARS-CoV-2 and antiviral-specific T-cell responses by IFNᵧ ELISpot assay. B. Intensity of positive SARS-CoV-2 specific immune responses in healthy donors and COVID-19 patients. Mann Whitney test, **P>0.001. Median with interquartile range were indicated. C. Heatmap showing the positivity or the negativity of the serology index, T-cell immune responses to SARS-CoV-S, M and N proteins for each patients included in the study (online Morpheus software). D. Frequency of patients with a specific T-cell response for at least one SARS-CoV-2 proteins (CoV-S, CoV-M or CoV-N) (P = 0.0211). E. Frequency of mild illness and severe pneumonia patients with specific T-cell responses for simultaneously CoV-S, CoV-M and CoV-N proteins (P = 0.0191). F. Frequency (%) of positive antiviral memory CD8T-cell responses for healthy donors, mild illness and severe pneumonia COVID-19 convalescent patients. Healthy donors were represented by light gray points. COVID-19 convalescent patients were respectively represented by black point (mild illness) and red points (severe pneumonia). PBMC: Peripheral Blood Mononuclear Cells.
Beyond specific cellular responses, typical humoral responses to acute viral infection are wildly induced in COVID-19 patients.8 Zhao et al. showed that the seroconversion rate and antibody levels increased rapidly during the first two weeks with a cumulative seropositive rate of 50.0% on the 11th-day and 100% on the 39th-day. High titers of IgG antibodies detected by Enzyme immunoassays have been shown to positively correlate with neutralizing antibodies. 8 , 9 In this study, the median serology index of severe pneumonia patients was equal to 7.19 S/CO [IQR: 6.1–8.28] and was significantly higher than the one of mild illness patients that was equal 4.92 S/CO [IQR: 2.52–7.32], (P<0.001) (Table 1). A heatmap was then generated, using the online Morpheus software (https://software.broadinstitute.org/morpheus/), to correlate all immunological parameters investigated (serology index and T-cell immune responses to SARS-CoV-S, M and N proteins) ( Fig. 1C). We observed that all patients with severe pneumonia had a positive serology index and most of them had at least one specific cellular response for SARS-CoV-2 proteins (28 out of 30). In contrast, patients with mild illness had less specific cellular responses (20 out of 29) than severe pneumonia patients (P = 0.0211) (Fig. 1D). Among mild illness patients, three had a negative serology index (Fig. 1C). Specific T-cell responses for S, M and N proteins were simultaneously shown for 70.0% of severe pneumonia patients while only for 37.9% of mild illness patients (P = 0.0191) (Fig. 1E). Of note, levels of T-cell responses were not influenced by previous exposition to a specific COVID-19 treatment (Lopinavir/ritonavir; Interferon-Beta-1A; Hydroxychloroquine and Remdesivir) (data not shown). We notice that despite a lower intensity of response in terms of INFᵧ secretion (Fig. 1B), patients with severe pneumonia had frequencies of responses clearly distinct from the one of mild illness patients.
Another immunological issue is the potential impact of SARS-CoV-2 virus mediated infection on pre-existing memory T-cell repertoire against common viruses. To address this issue, we concomitantly measured in all the COVID-19 patients the reactivity against common viruses (Cytomegalo-, Epstein-Barr and Flu-virus: CEF) using IFNᵧ ELISpot assay. Median intensities of CEF specific T-cell responses in healthy donors, mild illness and severe pneumonia patients were similar in terms of intensities (respectively 144.0 IFNᵧ spot/3 × 105 cells [IQR: 48.8–665.9], 88.2 IFNᵧ spot/3 × 105 cells [IQR: 42.8–218.6] and 116.0 IFNᵧ spot/3 × 105 cells [IQR: 67.5–179.0]) and frequencies (respectively 83.0%, 76.0% and 77.0%) (Fig. 1F).
The present study shows the existence of both specific SARS-CoV-2 cellular and humoral responses depending on COVID-19 severity. The presence of potent adaptive immunity even in patients who underwent severe pneumonia sustain the rationale for the development of protective therapeutics against SARS-CoV-2 virus.
Author contributions
Designing research studies: MK, LV, CB, SL, VW, SPF, GC, CC and KB conducting experiments: LS, AB, EO, QL, MD, LM, LB acquiring data: LS, AB, EO, GE analyzing data: LS, MK, QL, KB providing reagents: QL, LV, CB, CC, ALC, SL and writing the manuscript: MK, LS, CB, KB.
Declaration of Competing Interest
Authors declare no competing financial interests.
Acknowledgement
We thank Olivier Adotévi for scientific support and proof reading. We thank Sylvie Cour (nurse) of the clinical investigation center (INSERM CIC 1431) for her help to collect blood sample. We also thank the Biomonitoring platform (Eléonore Gravelin, Adeline Renaudin, Harmonie Simonin, Caroline Laheurte) for technical support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.08.036.
Appendix. Supplementary materials
References
- 1.Bo X., Cun-Yu F., An-Lu W., Yi-Long Z., Yi-Han Y., Cong H. Suppressed T cell-mediated immunity in patients with COVID-19: a clinical retrospective study in Wuhan, China. J Infect. 2020;81(1):e51–e60. doi: 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steven T., Michiel H., Hendrik G., van der Kieft R., Chantal R., Kristin K. Elevated nucleoprotein-induced interferon-γ release in COVID-19 patients detected in a SARS-CoV-2 enzyme-linked immunosorbent spot assay. J Infect. 2020;81(3):452–482. doi: 10.1016/j.jinf.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alba G., Daniela W., Ramirez Sydney I., Jose M., Jennifer M. D., Rydyznski M.C. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181(7) doi: 10.1016/j.cell.2020.05.015. 1489-1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guang C., Di W., Wei G., Yong C., Da H., Hongwu W. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. 2020Doi: 10.1172/JCI1372442020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolas V., Graham J. B., Conor G., Samarth H., Joel K., Maria K. Immunology of COVID-19: current state of the science. Immunity. 2020 doi: 10.1016/j.immuni.2020.05.002. S1074761320301837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faraz A.S., A. Q.A., Matthew R. M. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses. 2020;12(3) doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aiping W., Yousong P., Baoying H., Xiao D., Xianyue W., Peihua N. Genome composition and divergence of the novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juanjuan Z., Quan Y., Haiyan W., Wei L., Xuejiao L., Yingying S. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kai-Wang T.K., Tak-Yin T.O., Wai-Shing L., Raymond T.A., Tak-Chiu W., Christopher L.D. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.