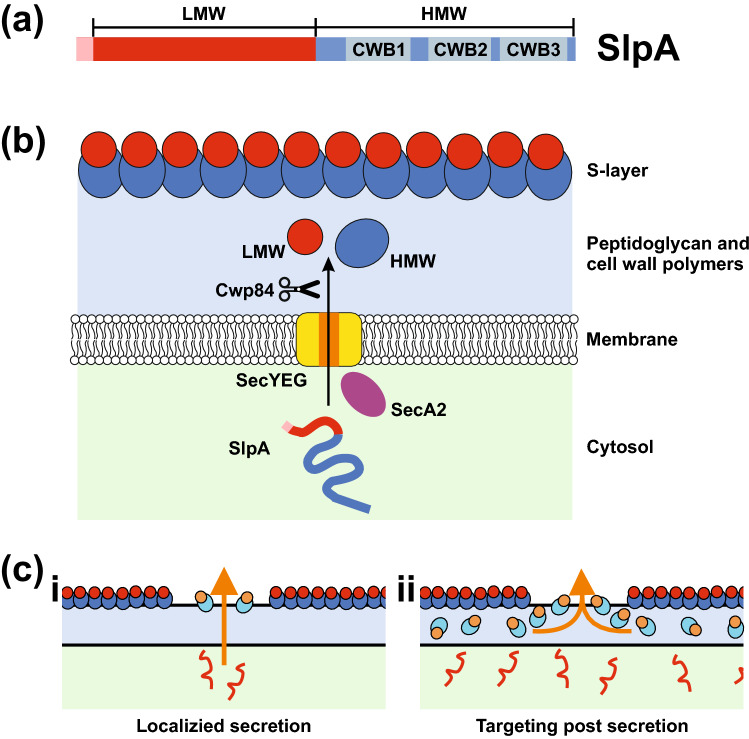
Figure 1.
The C. difficile S-layer and the SlpA secretory pathway. (a) Domain structure of SlpA precursor protein with signal sequence (Pink), low molecular weight region (LMW, Red) and high molecular weight region (HMW, Blue) that contains three cell wall binding domains (CWB, 1–3 in Grey). (b) Schematic diagram of SlpA secretion and processing in C. difficile. SlpA (Pink/Red/blue line) is translated in the cytosol (light green) and targeted for secretion across the membrane using SecA2 (Purple oval) most likely via the SecYEG Channel (Yellow/Orange). Cwp84 (Scissors) cleaves SlpA into low molecular weight (LMW, Red spheres) and high molecular weight (HMW, Blue spheres) S-layer protein (SLP) subunits. The HMW and LMW SLPs assemble to form hetero-dimers that incorporate into the S-layer. The surface of the S-layer consists largely of exposed LMW-SLP anchored to the cell wall (light blue) via cell wall biding domains of the HMW-SLP component (see (a)). (c) Models of SlpA integration into the S-layer. (i) unfolded SlpA (red line) is secreted from specific points on the cell membrane—directed by gaps in the S-layer or cell wall (colored as in (b)), newly processed SlpA (LMW-SLP orange circles, HMW-SLP light blue ovals) is transported directly through the cell wall for integration into the S-layer. Alternatively; (ii) SlpA is translocated across the cell membrane at multiple sites. A pool of SlpA lays within the cell wall ready to fill gaps in the S-layer.