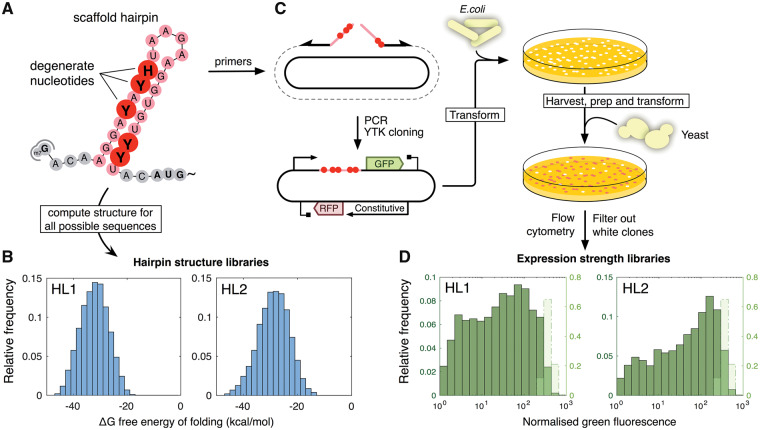
Figure 1.
Overview of 5′UTR hairpin library HL1 and HL2 creation. (A) Degenerate nucleotides are inserted at various positions into the design of a hairpin scaffold. (B) The MFE of all possible sequences is calculated with RNAfold and visualized in a histogram. These values are then used to predict the expression profile of the resultant library. If necessary, the composition and location of the degenerate nucleotides is adjusted to produce the required distribution of expression levels. (C) When the design meets the requirements, primers incorporating the required degeneracies are ordered. A library of plasmids for Escherichia coli transformation is created using PCR and a subsequent Golden Gate based assembly step implemented in YTK format. The library of E. coli transformants is harvested and plasmids prepped for yeast transformation. (D) Transformant yeast colonies are pooled and analyzed using flow cytometry. Clones that do not show constitutive red fluorescence are discarded in quality control for correct assembly. The diversity of the library of hairpins is reflected in the spread of green fluorescence over three orders of magnitude. For reference, the distribution of the original construct is shown in light green and normalized with a secondary axis. A pre-normalization comparison is shown in Supplementary Figure S2.