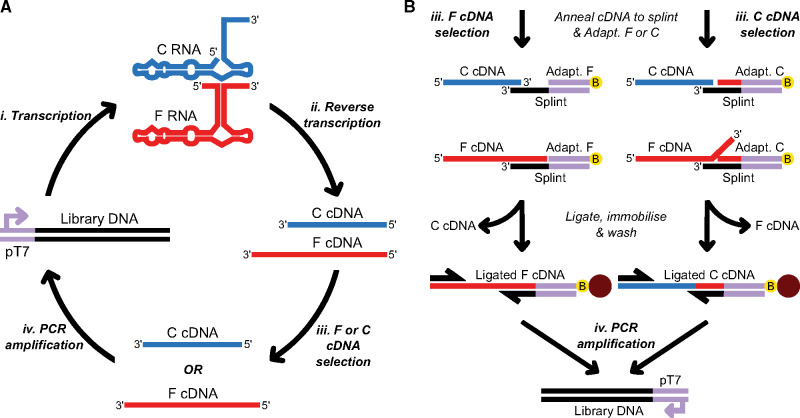
Figure 1.
LigASERR scheme. (A) During a single selection, Library DNA is transcribed yielding cleaved (abbreviated C) and full-length (abbreviated F) RNA, respectively (step i). RNA is reverse transcribed yielding cDNA (step ii). Either C or F cDNA is selected depending on the phenotype being enriched (step iii). Selected cDNA is PCR amplified (step iv). (B) Selection of full-length or cleaved cDNA (step iii) is initiated by annealing cDNA to a Splint with either Adapter (Adapt.) F or C, respectively. Only the desired cDNA forms a ligase substrate. Selected cDNA is ligated, ensuring sequences required for DNA template regeneration are incorporated. Additionally, the 3′ biotin modification present on each adapter facilitates immobilization and subsequent washing on streptavidin-coated paramagnetic beads, preparing the pool for amplification.