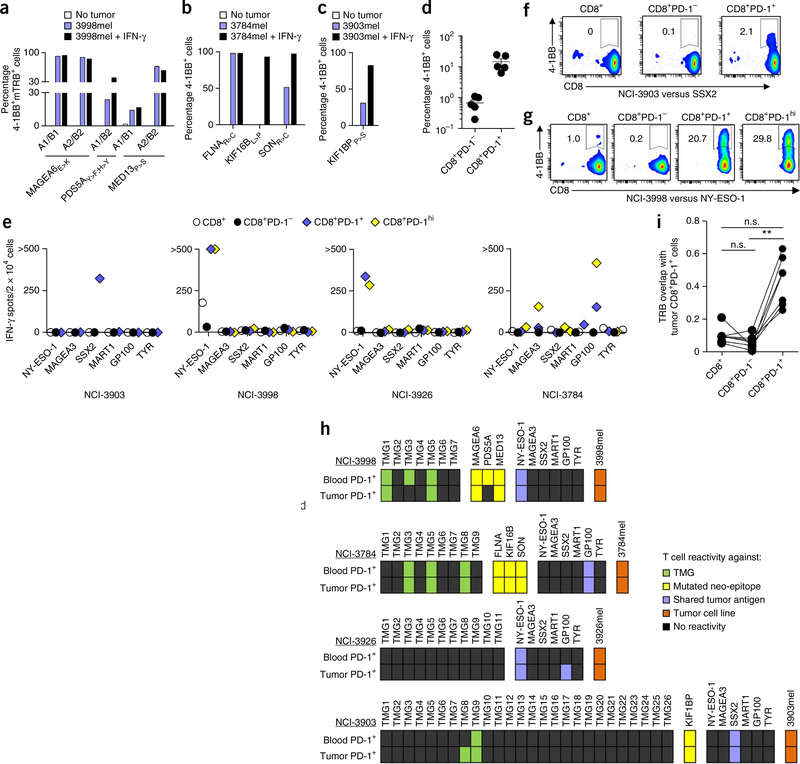
Figure 4.
Recognition of tumors and self-antigens by TCRs or CD8+ T cells isolated from peripheral blood, and comparison of the specificity and TCR repertoire between circulating and tumor-infiltrating CD8+ T cell subsets. (a–c) Reactivity (as determined by 4–1BB upregulation on CD3+CD8+ cells) of retrovirally transduced lymphocytes from subject NCI-3998 expressing MAGEA6E>K-, PDS5AY>F;H>Y- or MED13P>S-specific TCRs (a), circulating FLNAR>C-, KIF16BL>P- or SONR>C-specific lymphocytes from subject NCI-3784 (b) and KIF1BPP>S-specific lymphocytes from subject NCI-3903 (c) that were cocultured with their corresponding autologous tumor cell lines (3998mel, 3784mel and 3903mel, respectively) pretreated with or without IFN-γ. (d) Reactivity of the circulating CD8+PD-1− and CD8+PD-1+ lymphocytes from subjects NCI-3998, NCI-3784, NCI-3903, NCI-3926 and NCI-3713 to their corresponding autologous tumor cell line. Each dot represents the frequency of 4–1BB upregulation for one patient sample (n = 5). Mean ± s.e.m. is shown. (e–g) IFN-γ ELISPOT assays (e) and analysis of 4–1BB upregulation by flow cytometry (representative plots shown, gated on CD3+ cells) (f,g) of pretreatment PBMC CD8+ subsets from subjects (indicated below each graph) that were screened for recognition of the shared tumor antigens indicated. Experiments were performed without duplicates. All data are representative of at least two experiments. (h) Antigens recognized by circulating and tumor-infiltrating CD8+PD-1+ lymphocytes. Each rectangle represents a target antigen screened. (i) Deep-sequencing analysis of TRB from intratumoral CD8+ PD-1+ cells and matched pretreatment PBMC CD8+, CD8+PD-1− and CD8+PD-1+ cells (n = 7) was used to determine TRB sequence overlap between the tumor-resident CD8+PD-1+ cells and the blood-derived CD8+, CD8+PD-1− and CD8+PD-1+ cells (see Online Methods for calculation methodology). A TRB sequence overlap of 1 indicates 100% similarity between two populations. **P < 0.01 using Dunn’s test for multiple comparisons; n.s., not significant.