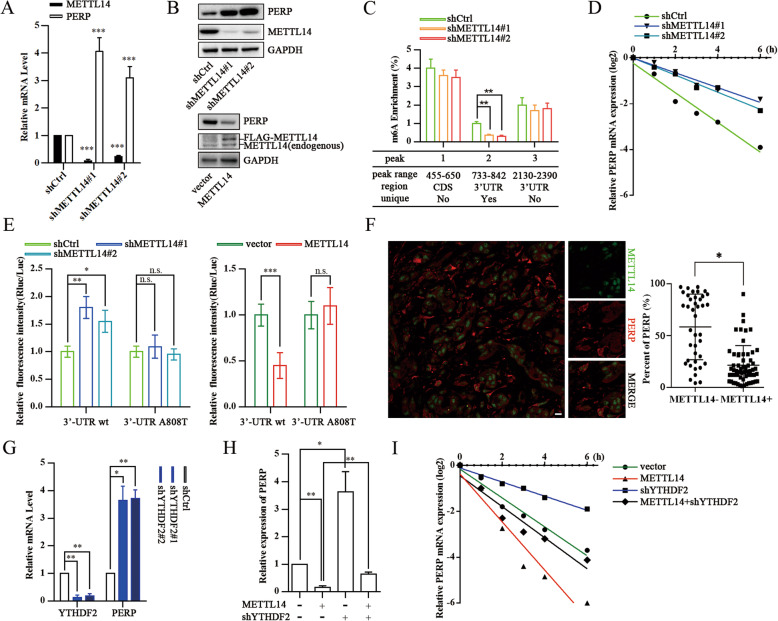
Fig. 5.
PERP is the key target of METTL14 in pancreatic cancer. a qPCR analysis of METTL14 and PERP in PANC-1 cells expressing shCtrl or shMETTL14. ***p < 0.001. b Western blotting of METTL14 and PERP in PANC-1 cells expressing shCtrl, shMETTL14, vector or METTL14. c MeRIP-qPCR analysis of fragmented PERP RNA from PANC-1 (control and METTL14 depleted) cells. ** p < 0.01. d qPCR analysis of PERP mRNA levels in PANC-1 cells (control and METTL14 depleted) after actinomycin D treatment. e PANC-1 cells were pre-transfected with wild-type or mutated pmiR-RB-Report-PERP-3′UTR plasmids, and then treated as indicated. Renilla luciferase activity was normalized to firefly luciferase activity and expressed as the mean ± SD. * p < 0.05; ** p < 0.01; ***p < 0.001; n.s., no significance. f Correlation between METTL14 and PERP protein levels in pancreatic cancer specimens. Left - representative IF images of pancreatic cancer specimens. Scale bar, 20 μm. Right - percentage of PERP-positive cells among METTL14-positive versus METTL14-negative cells in selected microscope fields of each tumor (compared by the t-test). * p < 0.05. g qPCR analysis of PERP in PANC-1 cells (control and YTHDF2 depleted). (H) qPCR analysis of PERP in PANC-1 cells (control and YTHDF2 depleted) in the absence or presence of METTL14 overexpression. * p < 0.05; ** p < 0.01. i qPCR analysis of PERP mRNA levels in PANC-1 cells (control and YTHDF2 depleted) in the absence or presence of METTL14 overexpression, and after actinomycin D treatment