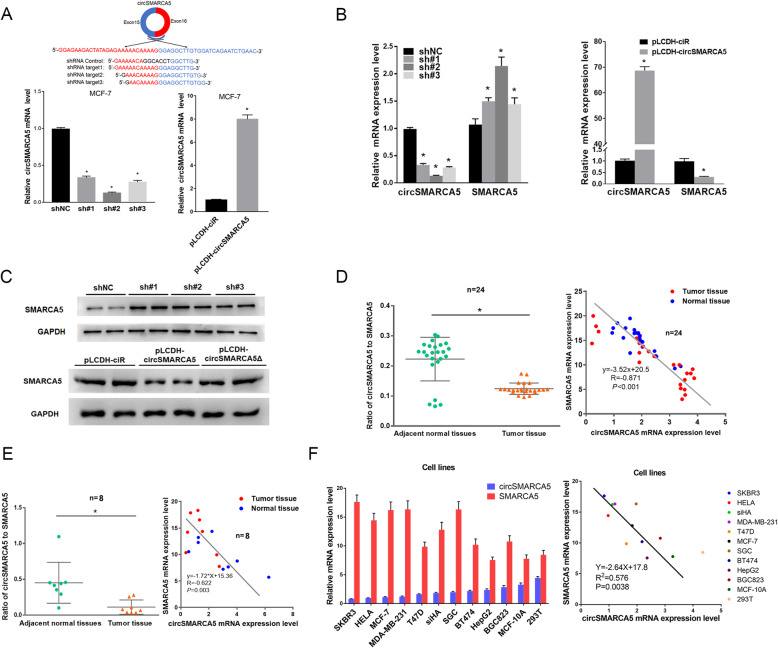
Fig. 2.
circSMARCA5 decreases the expression of SMARCA5 in cells. a Generation of circSMARCA5-knockdown and circSMARCA5-overexpressing cells. MCF-7 cells were infected with lentiviruses expressing shRNA against circSMARCA5 (sh-circSMARCA5; three different oligonucleotides) or circSMARCA5 (pLCDH-circSMARCA5). RT-qPCR was performed to evaluate the expression of circSMARCA5. GAPDH was used as an internal control. b RT-qPCR showing the levels of circSMARCA5 and SMARCA5 in MCF-7 cells stably expressing sh-NC, sh-circSMARCA5, pLCDH-ciR (control), or pLCDH-circSMARCA5. c Western blot showing the levels of SMARCA5 in MCF-7 cells stably expressing sh-NC, sh-circSMARCA5, pLCDH-ciR (control), pLCDH-circSMARCA5, pLCDH-circSMARCA5Δ (without splicing-inducing sequence). GAPDH was used as an internal control. (D-F) The ratio of circ-to-linear of circSMARCA5 in tumor tissue were significantly lower than normal tissue in breast cancer samples (d) and RCC samples (e) (P < 0.05). A negative correlation between circSMARCA5 and SMARCA5 expression was observed in breast cancer samples (d), RCC samples (e), and various cell lines (f)