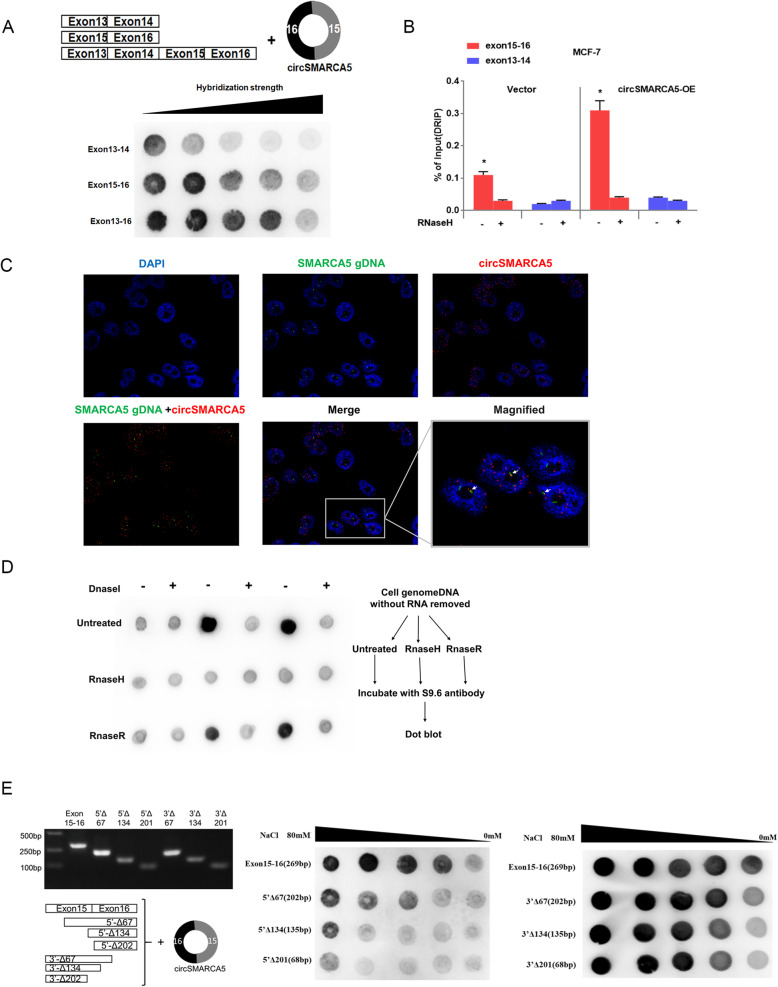
Fig. 5.
circSMARCA5 interacts with its site of transcription. a circSMARCA5 interacts with the 15-16 exon sequence of the SMARCA5 locus. A series of exon DNA fragments were hybridized with circSMARCA in vitro. The DNA-RNA hybridization strength was quantified by dot-blot with R-loop-specific S9.6 antibody. Hybridization stringency was altered by decreasing ionic strength (80-0.1 mM NaCl). b DRIP-qPCR analysis of the 15-16 exon sequence of SMARCA5 to detect the association of circSMARCA5 in MCF-7 cells. RNase H-treated and/or DRIP-qPCR analysis of the 15-16 exon sequence as a control. c CircSMARCA5 partially localized at its site of transcription. Double FISH of circSMARCA5 (red) and its parent DNA region (green). The nucleus was stained by DAPI. d Dot-blot of R-loops in MCF-7 cell genomic DNA preparations treated with DNase I, RNase H, or RNase R. The DNA-RNA hybrids in genome DNA were analyzed by S9.6 antibody. e Mapping of the R-loop formation region of circSMARCA5. A series of exon 15-16 deletion mutants were hybridized with circSMARCA5 for the dot-blotting assay. The DNA-RNA hybridization intensity was analyzed by dot-blot with an S9.6 antibody targeting the DNA-RNA hybrid strand. Hybridization stringency was altered by decreasing ionic strength (80-0.1 mM NaCl)