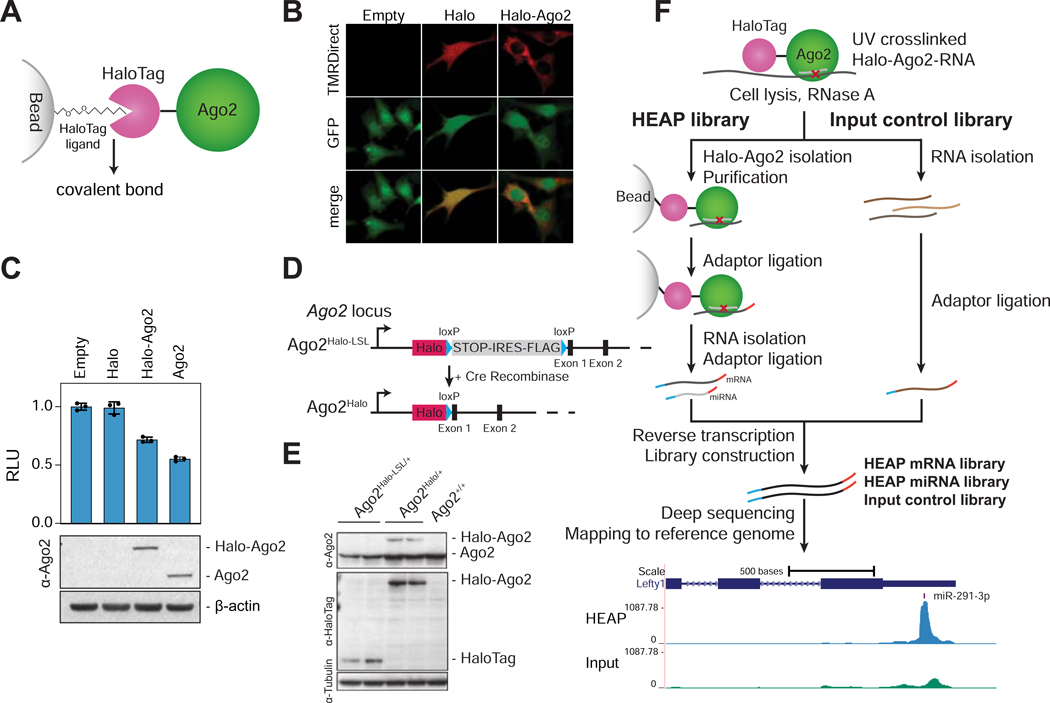
Figure 1. Halo-enhanced Ago2 pulldown (HEAP).
A) Schematic of the Halo-Ago2 fusion protein covalently bound to a bead-conjugated HaloTag ligand. B) Ago2−/− immortalized MEFs transduced with MSCV-PIG, MSCV-PIG-Halo or MSCV-PIGHalo-Ago2 retroviruses were incubated with the HaloTag TMRDirect ligand and imaged. Notice the prevalently cytoplasmic localization of the Halo-Ago2 fusion protein. C) Ago2−/− MEFs transduced with retroviral vectors encoding HaloTag alone, full length Ago2 or the Halo-Ago2 fusion protein were transiently transfected with reporter plasmids expressing Firefly and Renilla luciferase and a plasmid expressing an shRNA against the Firefly luciferase. The ratio between Firefly and Renilla luciferase activity was measured 48 hours after transfection (upper panel). Whole-cell lysates from the same cells were probed with antibodies against Ago2 and β-actin (lower panel). Error bars: Mean ± SD. D) Schematic of the targeting strategy used to generate the Halo-Ago2 conditional knock-in allele. Halo: HaloTag; STOP: stop codon; IRES: internal ribosome entry site. E) Whole-cell lysates from mESCs with the indicated genotypes were probed with antibodies against Ago2, HaloTag and Tubulin. F) Outline of the strategy used to generate HEAP and input control libraries (upper panel) and a representative Halo-Ago2 binding site identified in mESCs (lower panel).
See also Figure S1A, S1B and Data S1.