FIG 1.
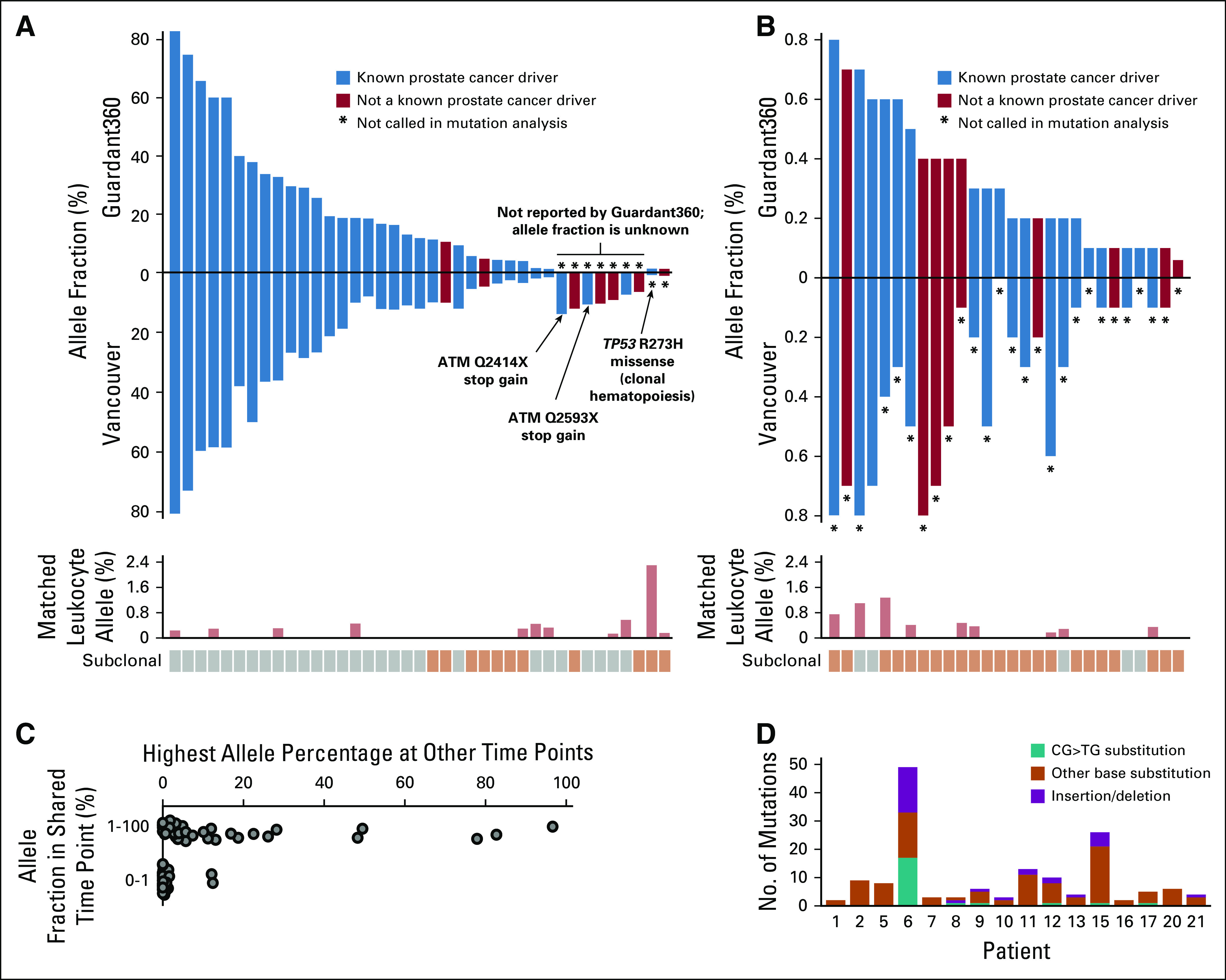
Concordance of somatic circulating tumor DNA mutation calls between Guardant360 and the Vancouver panel. Allele fractions of somatic mutations based on the two assays: mutations with allele fraction of (A) 1% or greater and (B) lower than 1%. Known prostate cancer driver mutations are shown in blue; other mutations in red. Bar plot below shows allele fraction in matched WBC samples. Mutations were labeled as subclonal if their allele fraction was less than half the allele fraction expected for truncal mutations (described in Patients and Methods). It is plausible that some mutations labeled as subclonal had a nonprostate origin. (C) Plot showing all mutations (dots) identified by Guardant360 in the cell-free DNA (cfDNA) time points that were also analyzed with the Vancouver panel, grouped by allele fraction. Position along x-axis indicates the highest allele fraction that those mutations reached in other time points analyzed by Guardant360. (D) Bar plot showing the total number of somatic mutations called by the Vancouver panel in 16 cfDNA samples with detectable mutations. Patient 6 displays a hypermutation signature enriched for somatic CG>TG transitions and insertions/deletions, consistent with an underlying mismatch repair defect.
