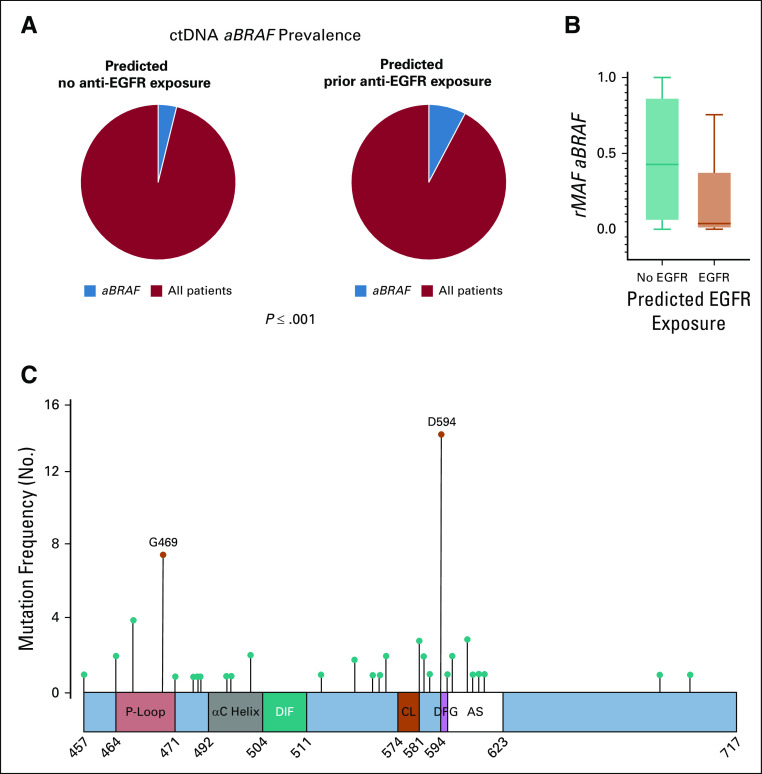
FIG 5.
Circulating tumor DNA (ctDNA) analysis of atypical, non-V600 BRAF (aBRAF) in cohort 2. (A) ctDNA prevalence of aBRAF (3.99%) among non-exposed patients (n = 2,880) v ctDNA prevalence of aBRAF (8.38%) among predicted anti-EGFR exposed (n = 644). (B) Relative mutational allele frequency (rMAF) of aBRAF among predicted anti-EGFR exposed v nonexposed patients, P < .001. (C) Frequency and location of mutations in the BRAF protein in mCRC patients predicted to be exposed to anti-EGFR antibodies. Schematic figure of the conserved region-3 (CR3) of the BRAF protein, also known as the BRAF kinase domain (amino acids 457 to 717), and the location of the class II and III hotspot mutations. The kinase domain contains the P-loop (amino acids 464-471), the αC helix (amino acids 492-504), the dimerization interface (DIF, amino acids 504-511), the catalytic loop (CL, amino acids 574-581), the DFG motif (amino acids 594-596), and the activation segment (AS, amino acids 594-623). class II, activating G469A mutations (P < .001) and class III, D594N (P < .001) were the most common, respectively, among predicted anti-EGFR exposed patients. Figure adapted.12 rMAF, relative mutational allele frequency.