Fig 5. RNAseq identifies differentially expressed genes in RxL21 compared to RxL21ΔEAR transgenic lines.
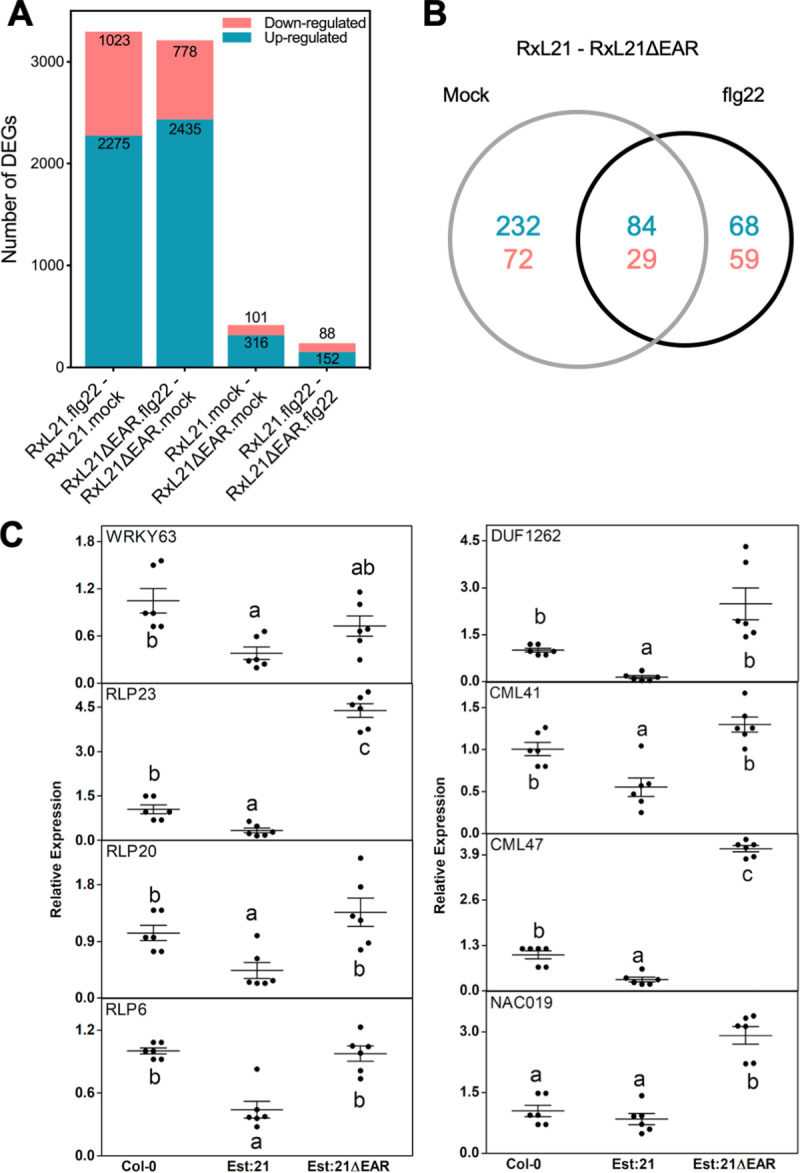
RNAseq was performed on HA-tagged RxL21 and RxL21ΔEAR-expressing transgenic lines. (A) The number of up- and down-regulated genes among differentially expressed genes under mock and flg22 treatment. DEGs were defined as having a log2 fold change ≥ 1 or ≤-1, and a BH adjusted p-value of < 0.05. (B) Venn diagram shows differentially expressed genes between RxL21 and RxL21ΔEAR after mock and flg22 treatment with an overlap of 84 upregulated and 29 down regulated genes. (C) Eight genes were selected for validation in estradiol (Est) inducible RxL21 and RxL21ΔEAR expressing lines and Col-0 WT. We treated Arabidopsis seedlings with 30 μM of β-estradiol for 18 hours. For Est:21 and Est:21ΔEAR, data were obtained from 2 independent lines each with 3 biological replicates and expression was normalised to Arabidopsis tubulin gene. Black circles are individual data points and bars denote the mean ± SE of target gene expression. Letters indicate significant differences (P < 0.05) (One-way ANOVA with Tukey’s honest significance difference).
