Abstract
The COVID-19 pandemic, caused by the novel coronavirus SARS-CoV-2, has led to several million confirmed cases and hundreds of thousands of deaths worldwide. To support the ongoing research and development of COVID-19 therapeutics, this report provides an overview of protein targets and corresponding potential drug candidates with bioassay and structure–activity relationship data found in the scientific literature and patents for COVID-19 or related virus infections. Highlighted are several sets of small molecules and biologics that act on specific targets, including 3CLpro, PLpro, RdRp, S-protein–ACE2 interaction, helicase/NTPase, TMPRSS2, and furin, which are involved in the viral life cycle or in other aspects of the disease pathophysiology. We hope this report will be valuable to the ongoing drug repurposing efforts and the discovery of new therapeutics with the potential for treating COVID-19.
Keywords: COVID-19, SARS-CoV-2, structure−activity relationship (SAR), bioassay, protein target, drug candidate
1. Introduction
COVID-19, the infectious disease caused by the newly emerged human coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2),1 was declared a global pandemic by the World Health Organization (WHO) on March 11, 2020. As of June 24, 2020, there have been more than 9 million confirmed cases and over 475,000 deaths worldwide.2 In order to combat this pandemic and prevent future recurrences, scientists around the world have been working tirelessly to elucidate the molecular basis for SARS-CoV-2 infection and to develop effective therapeutic agents and preventative vaccines.
SARS-CoV-2, a member of the Betacoronavirus genus, is an enveloped virus containing a single-stranded, positive-sense RNA genome.3 Two other members of this genus that also cause similar severe acute respiratory diseases in humans are severe acute respiratory syndrome coronavirus (SARS-CoV or SARS-CoV-1) and Middle East respiratory syndrome coronavirus (MERS-CoV). As RNA viruses, they use their RNA-dependent RNA polymerase (RdRp) to replicate their genomic RNA.4 In particular, SARS-CoV-2 and SARS-CoV share high levels of sequence homology and protein structural similarities.5 They both use cell membrane protein angiotensin-converting enzyme 2 (ACE2) as their receptor and need host serine proteinase TMPRSS2 to cleave or “prime” their spike (S) protein in order to fuse with the host cell membrane and enter the cells.6,7 Once inside the host cells, the viral genome is translated by the host cell protein synthesis machinery and the resulting polyproteins are autoproteolytically cleaved by coronavirus proteases 3-chymotrypsin-like protease (3CLpro) and papain-like protease (PLpro) to release smaller functional proteins to continue the viral replication process.8−11 In some cases, an excessive immune response called a cytokine storm may contribute to the further development of pulmonary edema, acute respiratory distress syndrome (ARSD), and systemic inflammation. Meanwhile, evidence is mounting that multiple organs may be damaged by SARS-CoV-2 infection in severe cases and that excessive blood coagulation may lead to life-threatening conditions.12−14
While there is no specific treatment for COVID-19, over the past few months, significant advances in discovering the molecular mechanisms of SARS-CoV-2 infection have been made, and numerous clinical trials have begun, with many more in the planning stages. For instance, several antiviral drugs or drug candidates already approved for other diseases, such as lopinavir/ritonavir and darunavir (anti-HIV), remdesivir (Ebola), chloroquine and its derivatives (antimalarial), as well as Arbidol and favipiravir (broad spectrum antiviral), were among the first drugs to be tested in multiple clinical trials across the world.15,16 Camostat mesilate and nafamostat, both TMPRSS2 inhibitors, were enlisted in clinical trials shortly after the indispensable function of TMPRSS2 in SARS-CoV-2 infection was discovered.7,17 Since the uncontrolled inflammatory response is one of the major contributing factors to disease severity, anti-inflammatory drugs, such as interferon β, baricitinib, tocilizumab, sarilumab, and acalabrutinib, are also being evaluated in clinical trials for usage either alone or in conjunction with another anti-SARS-CoV-2 agent.18−21 More recently, the potent anti-inflammatory effects of corticosteroids are being explored alone and/or in conjunction with other drugs. Initial reports showed that in patients hospitalized with COVID-19 dexamethasone resulted in a lower 28 day mortality among patients receiving respiratory support but not among those without respiratory support.22 In addition, several clinical trials are either underway or being planned that look at the effects of dexamethasone alone or in combination with other drugs.23
Although there are numerous ongoing clinical trials, only two drugs, remdesivir and favipiravir (avifavir), have so far been conditionally approved in a few countries for limited use,24−26 and these appear to show only modest effects. Moreover, the application of remdesivir is further limited because it can only be administered intravenously to hospitalized patients. Additionally, despite some conflicting results, multiple studies and meta-analyses have concluded that hydroxychloroquine offers either very small or no therapeutic effects in the treatment of COVID-19.27−29 The U.S. Food and Drug Administration (FDA) on June 15, 2020 revoked its Emergency Use Authorization for the use of hydroxychloroquine and chloroquine to treat COVID-1930 and issued cautions against its use due to the risk of incurring heart rhythm problems and other safety issues, including blood and lymph system disorders, kidney injuries, and liver damage and failure.31 However, clinical trials, including those on its use in prophylaxis, are continuing.29 As a result, there is still an urgent need to identify effective therapeutic agents for COVID-19 and possible future coronavirus-related diseases.
To support these efforts, we performed an analysis of the published journal articles and patents related to COVID-19 therapeutics. Herein, we review important viral and human targets and highlight target-based drug candidates with bioassay and structure–activity relationship (SAR) data from the Chemical Abstract Services (CAS)-indexed journal articles and patents.
2. Trends in COVID-19 Therapeutic Research
2.1. Journal Analysis
Since the beginning of the COVID-19 outbreak, the number of journal articles published on this topic has continued to increase as shown in Figure 1. Over the past 5 months, more than 16 000 articles have been published. This trend reflects the tremendous interest in the scientific community in understanding the new virus and in finding methods to combat the pandemic. A large number of publications are related to drug targets and the development of therapeutic agents. Due to the time requirements for de novo drug discovery, most efforts so far have focused on the repurposing of approved drugs, evaluation of investigational drugs (i.e., drugs in clinical trials for other purposes), molecular docking, and virtual screening studies.
Figure 1.
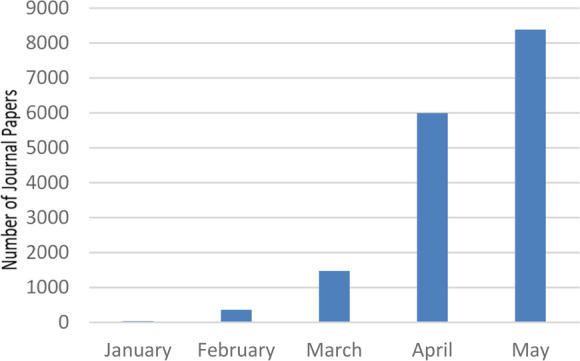
Monthly number of journal articles published related to COVID-19 in 2020.
Table 1 highlights some notable journal articles, published during this period, which focused on the development and identification of therapeutics against COVID-19. These were selected based on a number of factors, including journal impact factor, the number of citations/downloads, and the type of studies. Because our focus is on newly studied drug candidates, articles on well-known drugs such as remdesvir, hydroxychloroquine, and others that were presented in our earlier report are not included here.32 As shown in the table, both small molecules and biologics have been explored for the identification of COVID-19 therapeutics.
Table 1. Notable Journal Articles Related to COVID-19 Therapeutics.
| title | source | type of potential therapeutics | ref |
|---|---|---|---|
| Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors | Science (April 24, 2020) | small molecules (peptidomimetic α-ketoamides) | (59) |
| A human monoclonal antibody blocking SARS-CoV-2 infection | Nature Communications (May 4, 2020) | antibodies | (100) |
| A SARS-CoV-2 protein interaction map reveals targets for drug repurposing | Nature (April 30, 2020) | small molecules with different actions | (5) |
| An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice | Science Translational Medicine (April 29, 2020) | ribonucleoside analog that is also effective against CoV mutations resistant to remdesivir | (91) |
| Computational design of ACE2-based peptide inhibitors of SARS-CoV-2 | ACS Nano (April 14, 2020) | helical peptides that can be attached to nanoparticles and dendrimers | (501) |
| COVID-19: combining antiviral and anti-inflammatory treatments | Lancet Infectious Diseases (February 27, 2020) | baricitinib as an inhibitor of NAK family, especially for AAK1 | (502) |
| COVID-19: immunopathology and its implications for therapy | Nature Reviews Immunology (April 9, 2020) | biologics | (503) |
| Development of CRIPSR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza | Cell (April 29, 2020) | Cas13d-crRNAs | (504) |
| Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody | Nature (May 18, 2020) | human anti-SARS-CoV-2 S protein specific monoclonal antibody | (102) |
| Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2 | Cell (May 14, 2020) | human recombinant soluble ACE2 | (96) |
| Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2 | Cell Discovery (March 16, 2020) | various small molecules | (153) |
| Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds | Molecular Informatics (March 11, 2020) | 1000 potential ligands for SARS-CoV-2 3CLpro | (505) |
| Repurposing therapeutics for COVID-19: Supercomputer-based docking to the SARS-CoV-2 viral spike protein and viral spike protein-human ACE2 interface | ChemRxiv (February 27, 2020) | 48 potential small-molecule hits for S-protein–ACE2 interface and 30 hits for S protein alone | (506) |
| Therapeutic options for the 2019 novel coronavirus (2019-nCoV) | Nature Reviews Drug Discovery (February 10, 2020) | various small molecules and biologics | (8) |
| Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells | Cell (May 13, 2020) | neutralizing antibodies | (98) |
2.2. Patent Analysis
We also analyzed over 100 COVID-19-associated patents published in the first five months of 2020. These are categorized as follows: about 14% development of therapeutics including small molecules and biologics, 4% vaccines, 9% traditional Chinese medicines, and 56% the development of diagnostics.
Table 2 lists specific patent applications related to COVID-19 therapeutics. Patent application CN111135167A discloses that GC376 (CAS Registry Number (RN) 1416992–39–6), a 3CLpro inhibitor, significantly reduces SARS-CoV-2 replication in cells with an EC50 of 3.133 μM. Patent application CN111135166A discloses a pharmaceutical composition, consisting of GC376 and a prodrug, GS-441524 (CAS RN 1191237–69–0), which has a synergistic effect for inhibiting SARS-CoV-2 replication in cells with an EC50 of 1.0 μM. Due to the different examination processes at various patent offices, it is likely that a significant number of COVID-19 therapeutics related patents will be published later this year.
Table 2. Patents Related to COVID-19 Therapeutics.
| patent number | target | title | class |
|---|---|---|---|
| CN111184708A507 | 3CLpro | Application of silver monoethyl fumarate in resisting novel coronavirus infection | small molecules |
| CN111184707A508 | 3CLpro | Use of tolfenamic acid or a pharmaceutically acceptable salt thereof in the preparation of medicament for preventing and/or treating novel coronavirus inflammation | small molecules |
| CN111150833A509 | RdRp | Application of LTX-315 in preparing products for inhibiting coronavirus | small molecules |
| CN111135184A510 | RdRp | Application of GS-441524 in preparing novel coronavirus SARS-CoV-2 inhibitor | small molecules |
| CN111135167A511 | 3CLpro | Application of GC376 in preparing novel coronavirus SARS-CoV-2 inhibitor | small molecules |
| CN111135166A512 | RdRp, 3CLpro | Pharmaceutical composition consisting of GC376 and GS-441524 and application thereof in inhibiting novel coronavirus | small molecules |
| CN111053909A513 | 3CLpro, IL-6 | Application of 2019-nCoV 3CL hydrolase inhibitor and IL-6 monoclonal antibody in preparing medicament for treating coronavirus disease 2019 | small molecules |
| CN110960532A514 | RdRp | Composition containing benzylisoquinoline alkaloid and trans-resveratrol for treating coronavirus infection | small molecules |
| CN111166768A515 | ACE2 | Overexpression ACE2 mesenchyma cell in the preparation of medicine for treating new coronavirus application of drugs and preparation method thereof | biologics |
| CN111172195A516 | SARS-CoV-2, ORF1ab, nucleocapsid protein | Preparation method of gene therapy product for treating COVID-19 | biologics |
| CN111153991A517 | nucleocapsid protein | A human SARS-CoV-2 monoclonal antibody and preparation method and application thereof | biologics |
| CN111139242A518 | SARS-CoV-2 | Small-interfering nucleic acid, and its application for preparing pharmaceutical composition for preventing and/or treating new coronavirus pneumonia | biologics |
| CN111139241A519 | SARS-CoV-2 | Small interfering nucleic acid for inhibiting new coronavirus and its composition and application | biologics |
| KR2020032050520 | SARS-CoV-2 | COVID-19 virus customized triple knockout DNA treatment | biologics |
3. Key Proteins Involved in SARS-CoV-2 Infection
3.1. SARS-CoV-2 Proteins and Their Functions
SARS-CoV-2 contains a 30 kilobase long RNA genome, which encodes 16 nonstructural proteins (NSPs), 4 structural proteins, and 9 accessory proteins. The 16 NSPs are released by autoproteolysis of two large polyproteins by viral proteases, 3CLpro/NSP5 and PLpro/NSP3.5
Table 3 summarizes the functions of the SARS-CoV-2 proteins as well as their sequence similarities with those from SARS-CoV. The proteins are grouped based on their functions: (1) NSPs related to viral proteolysis, (2) NSPs related to viral RNA modification or polymerization, (3) structural proteins involved in viral particle assembly, and (4) accessory proteins with various functions. As indicated in the table, most of the SARS-CoV-2 proteins share high sequence similarities with those of SARS-CoV. So far, most attention has been focused on the S protein, 3CLpro/NSP5, PLpro/NSP3, and RdRp/NSP12 as potential drug targets. These proteins not only serve crucial functions in the viral lifecycle of SARS-CoV-2 but also have been well-studied in the related viruses SARS-CoV and MERS-CoV. Although less-studied, other proteins, such as NSP7/8/9/10/13/14/15/16, may also serve as drug targets. Conceivably, those that interfere with host immune regulation (e.g., NSP1 and Orf3b/6/9b) may also be potential targets for anticytokine storm drugs.5
Table 3. SARS-CoV-2 Proteins and Their Roles in Viral Infection.
| viral protein | role in SARS-CoV-2 infection | sequence similarity to SARS-CoV5 | |
|---|---|---|---|
| NSPs involved in proteolysis | NSP15,33 | inhibits production of proteins related to host innate immunity; overexpression increases production of pro-inflammatory chemokines | 91.1% |
| NSP25 | may serve as an adaptor for NSP3; not essential for viral replication | 82.9% | |
| PLpro/NSP35,33 | forms complex with NSP4 and NSP6; functions in stripping ubiquitin and blocking host innate immune response | 86.5% | |
| NSP433 | forms complex with NSP3 and NSP6; predicted to anchor replication complex to double membrane vesicles | 90.8% | |
| 3CLpro/NSP55,33,34 | cleaves polyproteins to release individual NSPs | 98.7% | |
| NSP65 | forms complex with NSP3 and NSP4; may also limit autophagosome expansion and lysosomal viral degradation | 94.8% | |
| NSPs involved in viral RNA modification and replication | primase/NSP75,34 | form primase complex as part of the replication complex (NSP7/8/12) capable of both de novo initiation and primer extension | 100% |
| primase/NSP85,34 | 99% | ||
| RNA-binding protein/NSP95 | single-stranded RNA-binding protein that interacts with replication complex (NSP7/8/12) | 98.2% | |
| NSP105,35 | zinc-finger protein that forms complex with NSP16 essential for replication; stimulates NSP16 to execute its methyltransferase activity; may also form complex with NSP14 to carry both exoribonuclease and methyltransferase activities | 99.3% | |
| RdRp/NSP125,34 | complexes with NSP7 and NSP8 to form RNA replication complex for viral replication and transcription | 98.3% | |
| helicase/NTPase/NSP135,34 | initiates the first step in viral mRNA capping; along with NSP14 and NSP16; installs the cap structure onto viral mRNA in the cytoplasm | 100% | |
| methyltransferase/exoribonuclease/NSP145,34 | corrects mutations during genome replication; facilitates capping of viral mRNA | 98.7% | |
| uridylate-specific endoribonuclease/NSP155,34 | essential for viral RNA synthesis | 95.7% | |
| 2′-O-methyltransferase/NSP165,34 | forms complex with NSP10; involved in mRNA S-adenosyl-l-methionine cap methylation | 98.0% | |
| NSP115 | short peptide with unknown function | 92.3% | |
| structural proteins | spike (S) protein5,7,36 | binds to ACE2 receptor on host cells and initiates viral fusion with host cell membrane | 87% |
| envelope (E) protein5,36 | plays a central role in viral morphogenesis and assembly | 96.1% | |
| membrane (M) protein5,36,37 | major driver for viral assembly | 96.4% | |
| nucleocapsid (N) protein5,36,37 | binds to viral RNA | 94.3% | |
| accessory proteins | ORF3a5 | involved in S protein trafficking and apoptosis | 85.1% |
| ORF3b5 | inhibits interferon activities | 9.5% | |
| ORF65,38 | interferon I antagonist that binds to karyopherins, alters their localization and reduces interferon/antiviral response | 85.7% | |
| ORF7a5 | involved in virus-induced apoptosis; inhibits CD317 which prevents release of coronavirus particles | 90.2% | |
| ORF7b5 | unknown function | 84.1% | |
| ORF85 | unknown function but not essential for virus replication | 45.3% | |
| ORF9b5 | involved in degradation of MAVS signalosome and limits host cell interferon responses | 84.7% | |
| ORF9c5 | unknown, may not be expressed | 78.1% | |
| ORF105 | unknown, may not be expressed | N.A. |
3.2. Human Proteins Involved in SARS-CoV-2 Infection
Similar to other viruses, SARS-CoV-2 not only relies on its own proteins but also utilizes many proteins from host cells to achieve its attack on the host cells. These host proteins may also be potential drug targets, since they play crucial roles in one or more aspects of the disease, as shown in Tables 4 and S1. The proteins in Table 4 are grouped into very broad categories such as viral entrance, viral RNA/protein synthesis, host inflammatory response, and other functions. As an example of the human proteins involved in virus entrance, ACE2 functions as the main receptor for the S protein of SARS-CoV-2,7 although other membrane proteins such as CD147/basigin may also be involved.39,40 After binding to a host cell receptor, the S protein needs to be cleaved by human proteases such as TMPRSS2, furin, or endosomal cathepsin L (CTSL) to initiate membrane fusion.41 Additional host proteins involved in various steps of SARS-CoV-2 infection and abnormal host responses such as cytokine storm-mediated inflammation and excessive blood clotting42 are also listed in Tables 4 and S1.
Table 4. Selected Human Proteins Involved in SARS-CoV-2 Infection.
| protein target | acronym | role in SARS-CoV-2 infection | COVID-19 clinical trial?a |
|---|---|---|---|
| Host Cell Proteins Involved in Viral Entrance | |||
| angiotensin-converting enzyme 243 | ACE2 | cell surface receptor for S protein | yes |
| furin44,45 | FURIN | cleaves S protein to expose S2 domain needed for virus-plasma membrane fusion | no |
| transmembrane serine proteinase 27 | TMPRSS2 | cleaves S protein to expose S2 domain needed for virus-plasma membrane fusion | yes |
| CD147/basigin39,40 | BSG | alternative cell surface receptor for S protein | yes |
| cathepsin L7,41 | CTSL | cleaves S protein to expose S2 domain needed for virus-endosomal membrane fusion | no |
| phosphatidylinositol inositol kinase PIKfyve41 | PIKFYVE | involved in phosphoinositide metabolism, regulates endosomal dynamics; may be involved in facilitating SARS-CoV-2 entry | no |
| Host Proteins Involved in Viral RNA/Protein Synthesis Processes | |||
| inosine monophosphate dehydrogenase IMPDH25,52 | IMPDH2 | binds to NSP14; involved in guanine nucleotide metabolism | no |
| translation initiation factor eIF-4A5,53 | EIF4A1 | binds to NSP2; involved in viral protein translation | no |
| translation initiation factor eIF-1A5 | EIF1AX | involved in viral protein translation | yes |
| translocon protein Sec615,54 | SEC61A, SEC61B | involved in viral protein insertion into endoplasmic reticulum | no |
| splicing factor SF3B155 | SF3B1 | altered expression in SARS-CoV-2 infection | no |
| Host Proteins Involved in Inflammatory Response | |||
| protein kinase JAK1 and JAK246,47 | JAK1/JAK2 | involved in cytokine signaling | yes |
| interleukin 648 | IL6 | involved in cytokine storm | yes |
| complement C349 | C3 | early complement factor that mediates inflammation and lung injury in COVID-19 | yes |
| chemokine receptor CCR115 | CCR1 | involved in cytokine signaling | no |
| chemokine CXCL1050 | CXCL10 | involved in cytokine signaling | |
| neutrophil extracellular traps42,51 | N.A. | increased levels in COVID-19 patients, excessive amounts trigger inflammation and blood clotting | yes |
Clinical trial data was obtained as of 5/21/2020 from www.ClinicalTrials.gov. “Yes” in the table includes trials with the following status: “Not Yet Recruiting”, “Recruiting”, “Enrolling”, “Active”, or “Completed”.
4. Distribution of Documents Associated with SARS, MERS, and COVID-19 and Therapeutic Substances Related to Specific Targets
Sequence homology between SARS-CoV-2 and SARS-CoV indicates that their key enzymes and structural proteins have high similarity.5 Molecular docking studies have revealed that antiviral agents effective against MERS-CoV and SARS-CoV may have similar affinities to the binding pockets in SARS-CoV2.56 Repositioning of existing therapeutic candidates developed for SARS and MERS has become a common theme during the past few months in the development of anti-COVID-19 therapeutics. As a result, many studies have explored the effects of potential drug substances initially developed or tested to combat other coronavirus infections. To help facilitate the ongoing repurposing efforts, we analyzed information published from 2003 to May 2020 related to SARS, MERS, and COVID-19 using the CAS content collection.
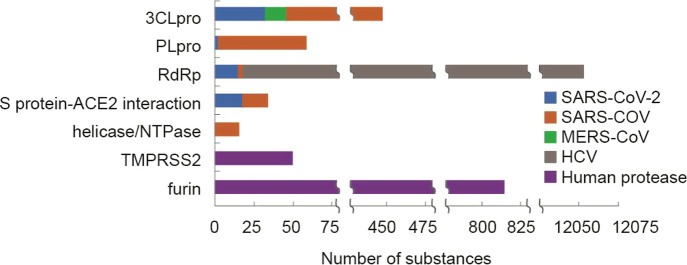
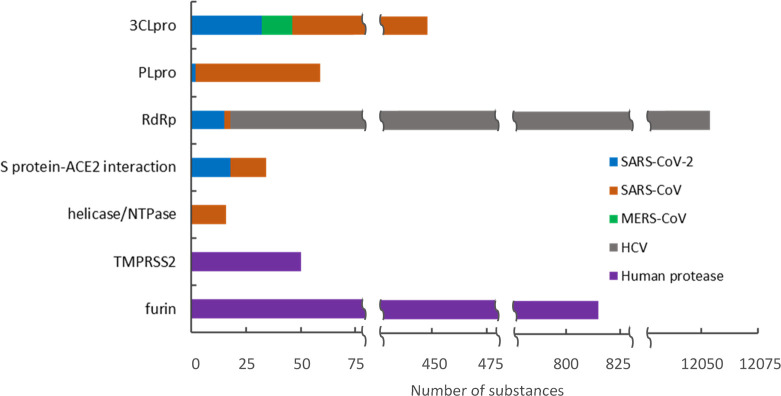
Of the 20,000 journal articles and 2,200 patents found in our analysis, over 500 patents and more than 500 journal articles were identified that contain potential therapeutic substances against SARS-CoV, MERS-CoV, and SARS-CoV-2 infections. The associations of potential therapeutic substances with specific targets in these documents were determined by data mining of CAS-provided index entries followed by intellectual review of the results. Figure 2 shows some high-frequency document–potential therapeutic substance–protein target relationships. The protein targets are listed according to the number of documents associated with each target. As shown in the figure, the 20 targets include the structural proteins (S and N proteins), nonstructural proteins (3CLpro, RdRp, PLpro, and helicase/NTPase), and human host proteins (ACE2, DPP4, TMPRSS2, and furin). Most of these studies appeared to have focused on the identification and development of small molecule therapeutics, but some biologics (biosequences) have also been developed, including some targeting the S and N proteins. Detailed information about some selected anti-SARS-CoV-2, SARS-CoV, and MERS-CoV substances will be discussed in the subsequent section.
Figure 2.
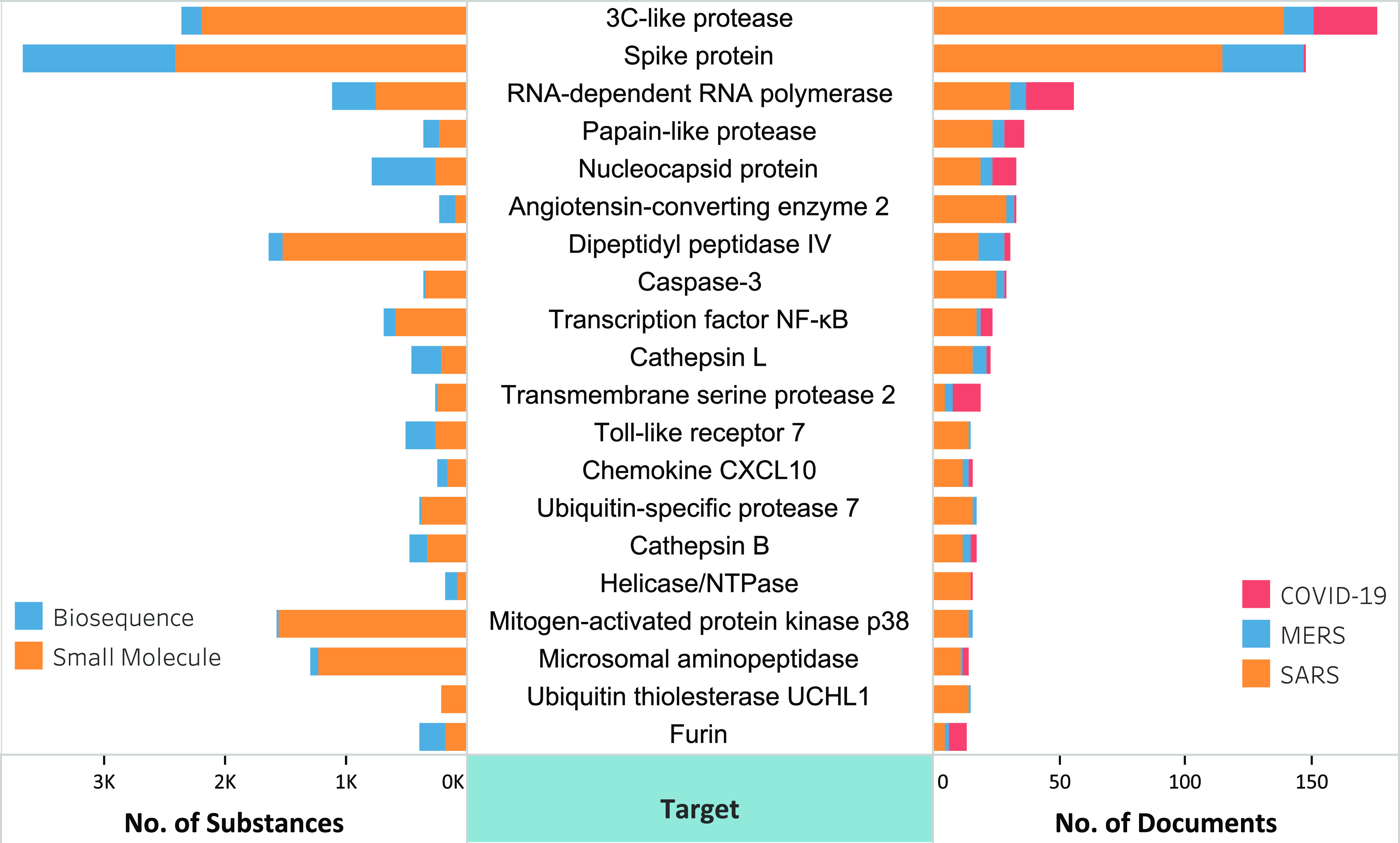
Distribution of SARS-, MERS-, and COVID-19-associated documents and potential therapeutic substances in relation to specific targets.
5. Bioassay and Structure–Activity Relationship Data for Small Molecules and Biologics against COVID-19 and Related Coronavirus Infections
5.1. Data Sources
In order to identify drug candidates for COVID-19, we extracted SARS-CoV-2-associated bioassay data related to the development of therapeutics from recently published journals. We also examined bioassay data related to human coronaviruses published in journals and patents from 2000 to 2019, which contain substance information, targets, activity measures [half maximal inhibitory concentration (IC50), half maximal effective concentration (EC50), inhibition constant (Ki), and dissociation constant (Kd)], and assay details. In this section, we focus on five viral proteins, 3CLpro, PLpro, RdRp, helicase/NTPase, and S protein, and two human proteases, TMPRSS2 and furin, that play a key role in S-protein-mediated cell entry of the virus. Selected substances with bioassay information toward these targets are presented in Tables 5–11 and in the Supporting Information. A high level view of the numbers of these substances associated with each protein target is found in Figure 3.
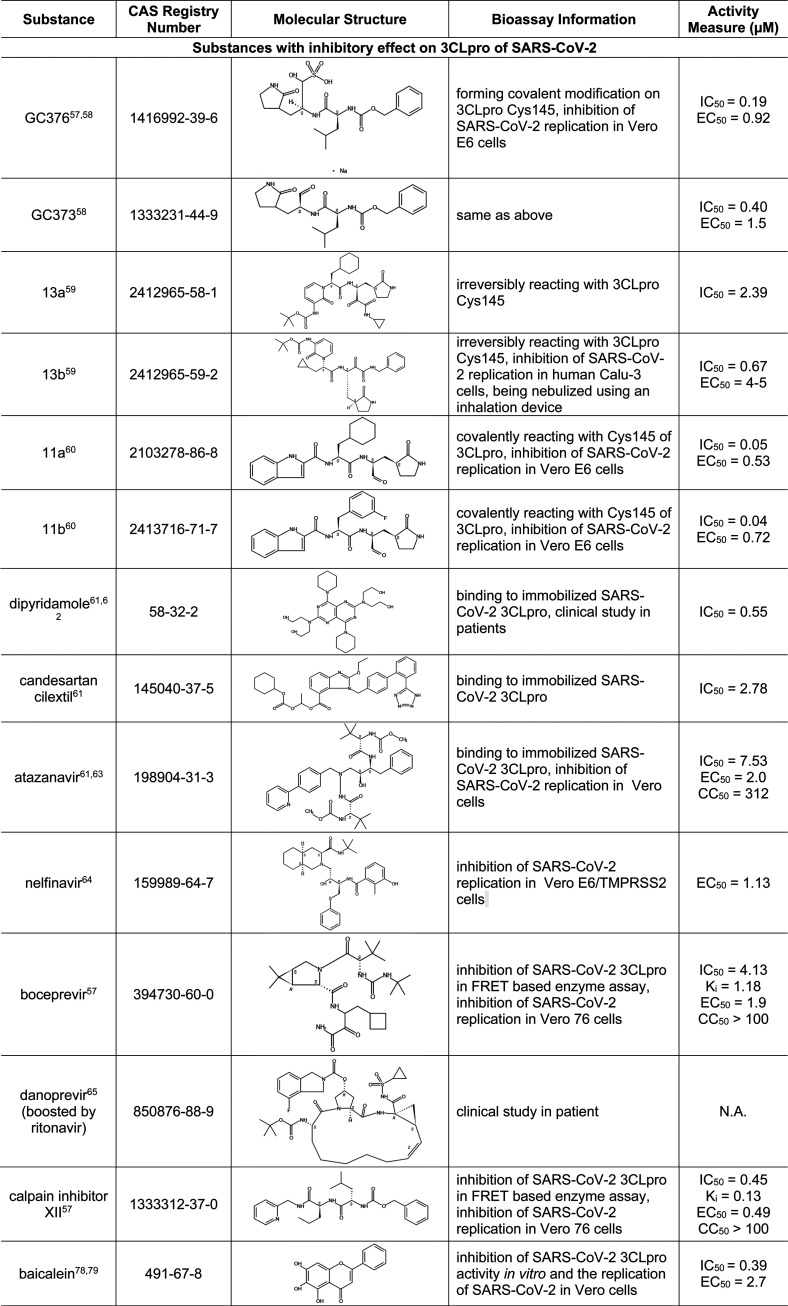
Table 5. Small-Molecule Inhibitors of 3CLpro in SARS-CoV-2 or SARS-CoV15,57−69,77−79.
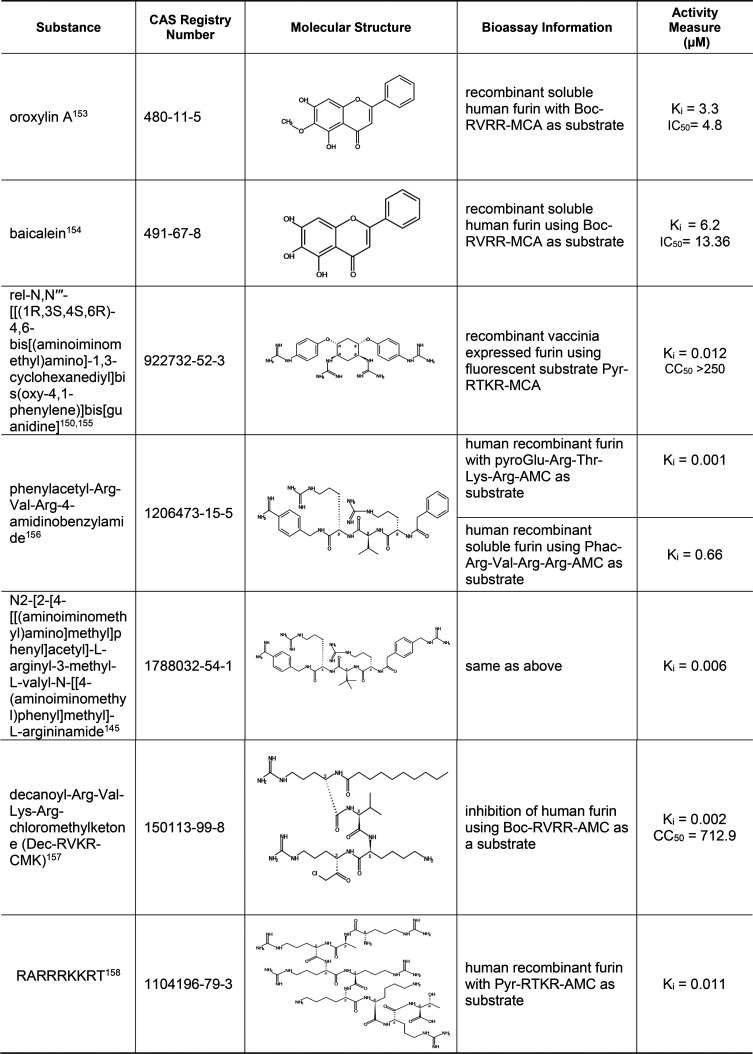
Table 11. Small-Molecule and Peptide Inhibitors of Human Protease Furin125,133−138.
Figure 3.
Numbers of substances with bioassay information for specific viral and human protein targets related to COVID-19 or other related viral infections.
5.2. Small-Molecule Inhibitors of 3CLpro
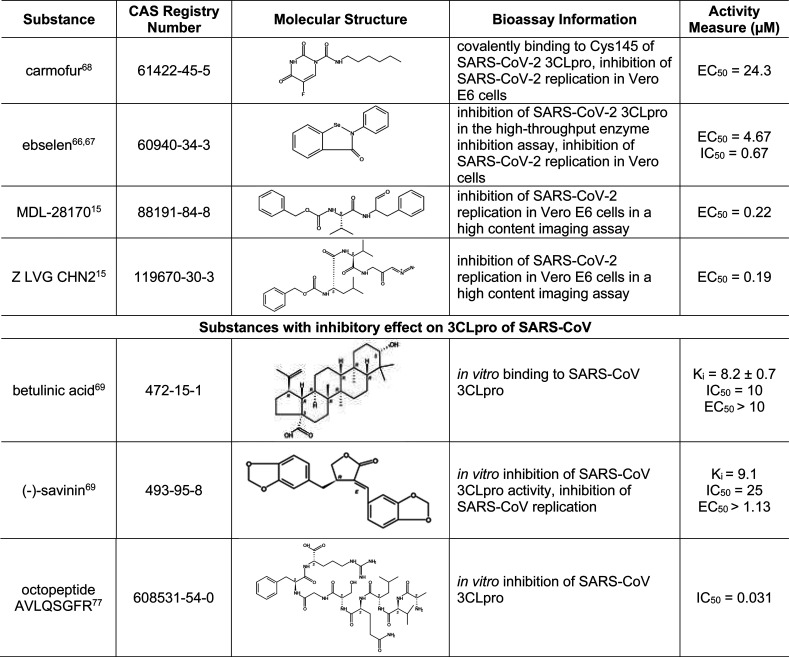
Of all the SARS-CoV-2 proteins, 3CLpro has the richest history of research data from other coronaviruses. Since 3CLpro is highly conserved among SARS-CoV-2, SARS-CoV, MERS-CoV, and other coronaviruses, previous research on this enzyme can serve as an excellent foundation for drug design of inhibitors of SARS-CoV-2 3CLpro. Table 5 highlights some substances that are active against 3CLpro of SARS-CoV-2 or SARS-CoV. Compounds GC376 and GC373 were designed based on the structures of 3CLpro from other viruses, but these were later shown to be also effective against SARS-CoV-2.57,58 Compounds 11a, 11b, 13a, and 13b were designed based on the recently revealed SARS-CoV-2 3CLpro crystal structure.59,60 In particular, 13a and 13b displayed longer plasma half-lives, and 13b can be nebulized for potential inhalant formulation.59 As can be seen in the table, all these compounds share a common pyrrolidinyl structure. Some substances in Table 5 were initially identified in computer-based predictive modeling studies, which have greatly expedited the identification of potential 3CLpro inhibitors. For example, Li et al. performed molecular docking studies followed by free energy perturbation (FEP) calculations of FDA-approved drugs and identified 25 drugs, which were further evaluated for their effect on SARS-CoV-2 3CLpro. Out of these 25 drugs, 15 displayed significant inhibitory activity against SARS-CoV-2 3CLpro.61 Shown in Table 5 are three such drugs with relatively low IC50 values, including dipyridamole, an anticoagulant currently in COVID-19 clinical trial,62 and atazanavir, an HIV protease inhibitor with both anti-3CLpro and anti-inflammation activities.63 Other clinically available protease inhibitors for other viruses, such as nelfinavir,64 boceprevir,57 and danoprevir,65 were also found to be potent inhibitors of 3CLpro. In particular, danoprevir boosted by ritonavir showed promising results in COVID-19 patients.65 Ebselen, an investigational drug with anti-inflammatory, antioxidant, and cytoprotective activities, has also been identified as 3CLpro inhibitor for SAS-CoV-2.66,67 Other drug candidates that functioned as cysteine protease inhibitors and inhibited SARS-CoV-2 infection include MDL-28170 and Z LVG CHN2, as identified from a large-scale drug repositioning screening of 12 000 FDA-approved and investigational drugs.15 Carmofur, an antineoplastic drug, covalently binds to 3CLpro Cys145 (a critical residue in the catalytic site) and inhibits viral replication in Vero E6 cells.68
3CLpro inhibitors discovered from SARS-CoV and MERS-CoV studies were also examined. A few of these compounds are shown in Table 5, and a more complete list is given in Table S2. Of these, both betulinic acid and savinin not only are 3CLpro inhibitors of SARS-CoV but also may act on other targets, with betulinic acid acting as a cannabinoid receptor (CB) modulator (CB1 antagonist/CB2 agonist) and savinin acting as a tumor necrosis factor α (TNF-α) antagonist.69−72 Activation of cannabinoid receptor 2 (CB2), mainly expressed in immune cells, is reportedly linked to inhibition of inflammation and cytokine storms.73,74 Thus, activation of CB2 and inhibition of TNF-α would lead to attenuation of cytokine storm commonly observed in severe cases of COVID-19. While seemingly attractive as potential drug candidates for COVID-19, the polypharmacological properties of betulinic acid (inhibition of 3CLpro and activation of CB2) and savinin (inhibition of both 3CLpro and TNFα) remain to be confirmed.75 In addition, several oligopeptides with or without chemical modification have been identified as 3CLpro inhibitors. For example, the octapeptide AVLQSGFR inhibited SARS-CoV 3CLpro and yet exhibited no cytotoxicity in Vero cells, indicating its potential as a drug candidate with low toxicity.76,77
5.3. Small-Molecule Inhibitors of PLpro
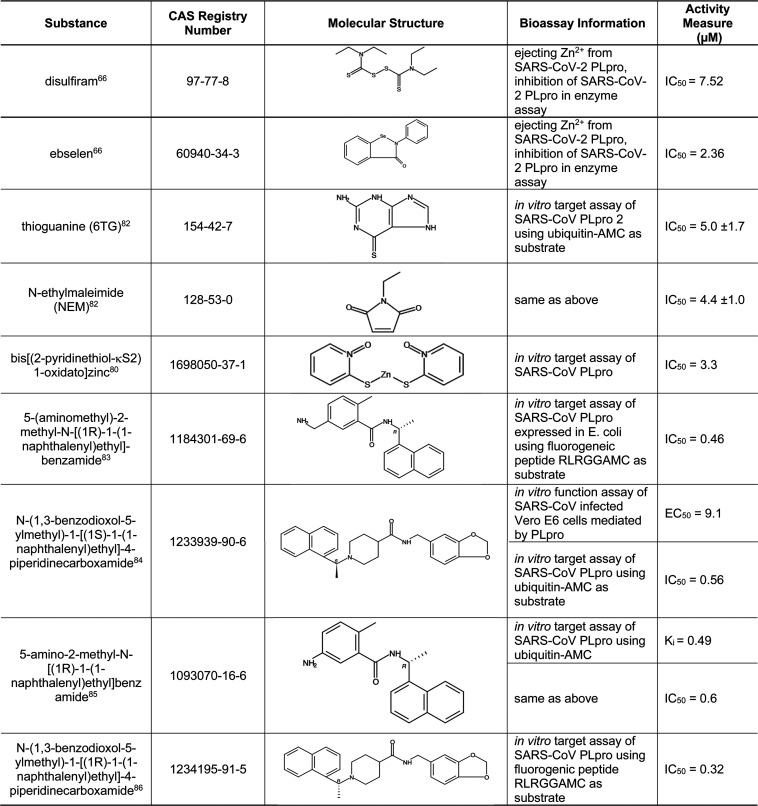
Besides its protease activity essential for viral replication, PLpro has the additional function of stripping ubiquitin and ISG15 from host-cell proteins to aid coronaviruses in escaping the host innate immune response. Therefore, inhibiting PLpro may be of use in not only inhibiting viral replication but also preventing the inhibition of innate immunity.10Table 6 presents selected small molecules shown to inhibit PLpro from SARS-CoV-2 or SARS-CoV. In a study with SARS-CoV-2 PLpro, two clinically safe Zn2+ ejectors, disulfiram and ebselen66 (also inhibit 3CLpro as shown in Table 5), were shown to extract Zn2+ from the critical cysteine residues of PLpro and inhibit its enzyme activity. Tioguanine, also known as 6-thioguanine (6-TG), is a chemotherapy agent that is on the World Health Organization’s List of Essential Medicines81 and could potentially be used to treat COVID-19. A more complete list of substances active against PLpro can be found in Table S3.
Table 6. Small-Molecule Inhibitors of PLpro66,80,82−86.
5.4. Small-Molecule Inhibitors of RdRp
Recently, the cryo-EM structure of RdRp has been revealed,87 which will undoubtedly help to guide the design of its inhibitors. Ideal RdRp inhibitors will not only terminate RNA synthesis catalyzed by RdRp but also have the potential to block its exonucleolytic proofreading activity. Because of these factors, RdRp inhibitors are often nucleotide analogs with modifications on the sugar or base.
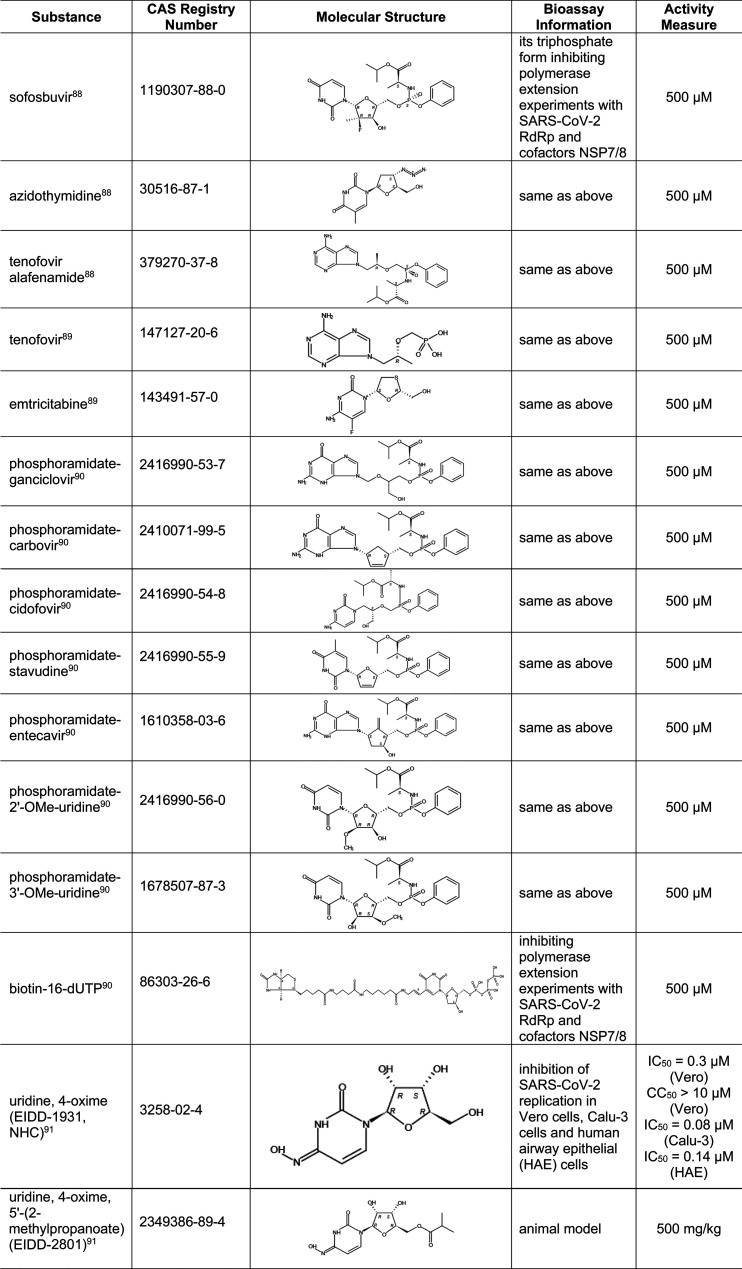
Table 7 provides compounds recently identified as inhibitors of SARS-CoV-2 RdRp in various bioassays. Included in this table are some FDA-approved drugs, such as sofosbuvir (a key component of hepatitis C drug EPCLUSA), azidothymidine (an anti-HIV drug), tenofovir alafenamide (a drug for HIV and hepatitis B), and tenofovir and emtricitabine (two components in DESCOVY and TRUVADA, two anti-HIV drugs).88,89 In addition, previously discovered SARS-CoV RdRp inhibitor EIDD-1931 was tested in SARS-CoV-2 and exhibited a high potency for infection inhibition.91 Its oral form, EIDD-2801, was also tested in animal models.91 A complete list of substances active against other (+)ssRNA viruses is shown in Table S4.
Table 7. Small-Molecule Inhibitors of RdRp in SARS-CoV-288−91.
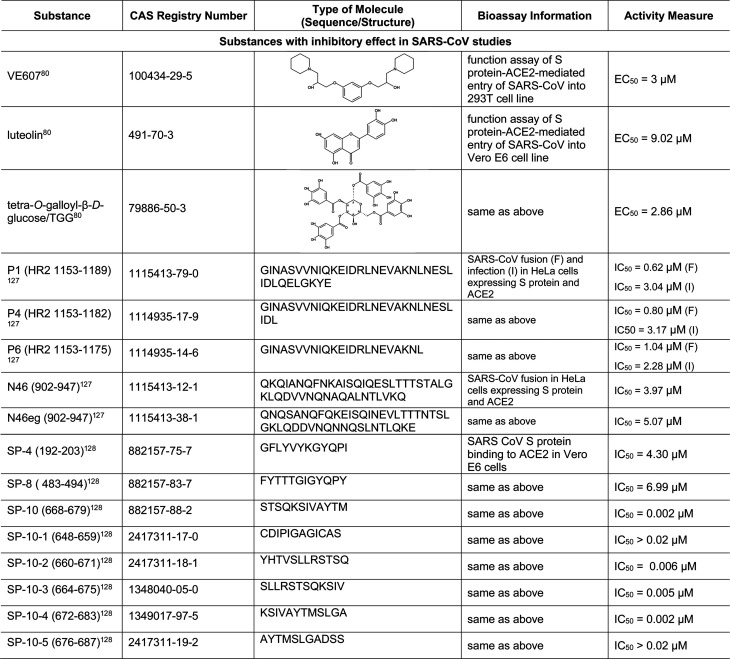
5.5. Small Molecules and Biologics That Affect Viral Entry Mediated by S-Protein–ACE2 Interactions
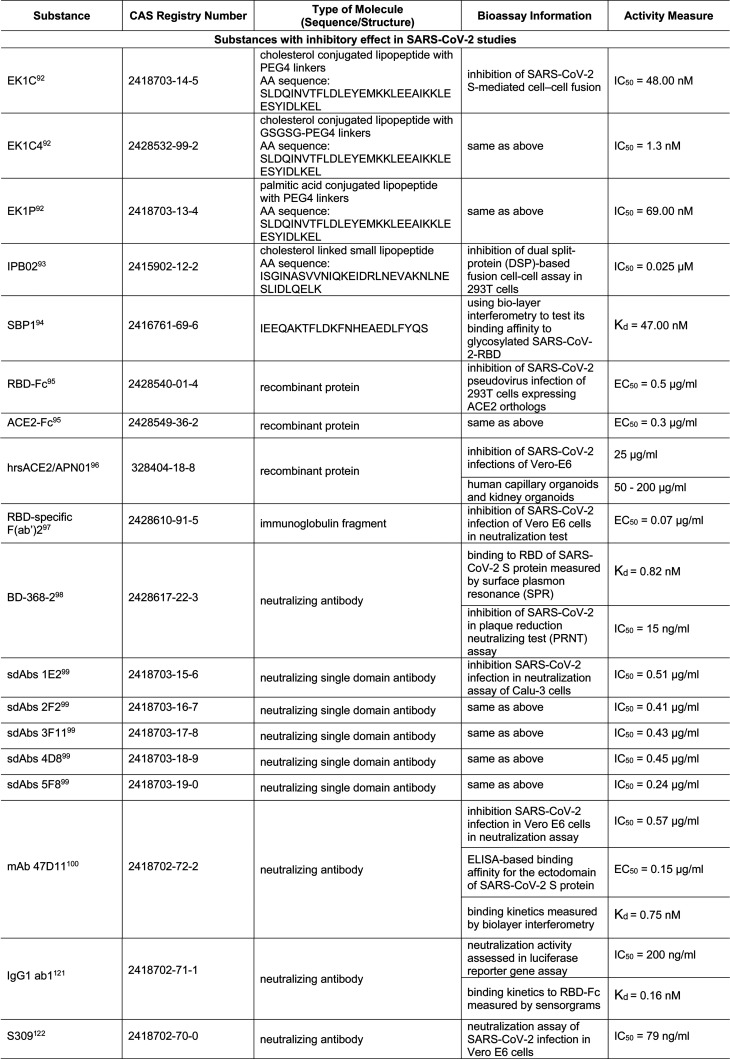
Unlike the viral proteases and RdRp, which are more likely to be inhibited by small molecules, inhibitors of the interaction of S protein with receptor ACE2 are predominantly small peptides and recombinant proteins mimicking ACE2 or neutralizing antibodies against the S protein. Recently, many of these biological molecules have been tested with SARS-CoV-2, as shown in Table 8. For example, when EK1, a peptidic pan-coronavirus fusion inhibitor which targets the heptad repeat (HR)1 region of the S protein, was linked to cholesterol (EK1C and EK1C4) or palmitic acid (EK1P), they displayed more potent inhibition against SARS-CoV-2 S-protein-mediated membrane fusion.92 Another lipopeptide, IPB02, is designed based on HR2 sequence and also showed strong activity in inhibiting the SARS-CoV-2 S-protein-mediated viral–cell fusion.93 SBP1, derived from the α1 helix of ACE2 peptidase domain, showed high affinity to the SARS-CoV-2-RBD.94
Table 8. Small Molecules and Biologics That Affect Viral Entry Mediated by S-Protein–ACE2 Interactions80,92−100,101,102,107,108.
Recombinant proteins ACE2-Fc and hrsACE2, which act as decoy receptors, also target the S-protein–ACE2 interaction and viral–host-cell membrane fusion.95,96 Furthermore, an increasing number of antibodies, immunoglobulin fragments, or even single-domain antibodies are being developed for this purpose, and their activities have been demonstrated in various assays.97−100,101,102
In addition to the inhibitors mentioned above that have been tested with SARS-CoV-2, we found more compounds from SARS-CoV experiments that could be valuable for SARS-CoV-2 treatment. For example, the small molecule VE607 inhibits both S-protein–ACE2 interaction-mediated SARS-CoV entry and SARS-CoV plaque formation.103 The flavonoid luteolin has been reported to bind to the S protein and inhibit SARS-CoV entry into host cells.104,105 It also has anti-inflammatory and 3CLpro inhibition activities.106
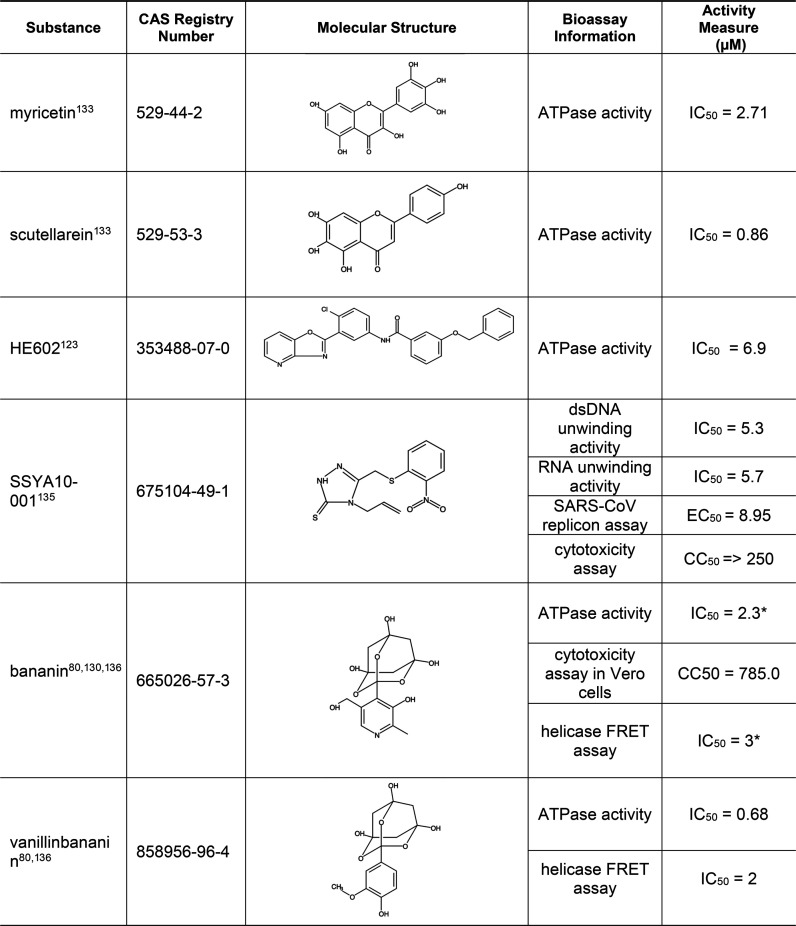
5.6. Small-Molecule Inhibitors of SARS-CoV Helicase/NTPase
As mentioned earlier in this report, NSP13 displays both helicase and NTPase activities and initiates the first step in viral mRNA capping. As part of a complex with NSP14 and NSP16, it installs the cap structure onto viral RNA in the cytoplasm. Since the sequence of SARS-CoV-2 helicase/NTPase is almost identical (100% in sequence similarity) to that of SARS-CoV,109 inhibitors of SARS-CoV helicase/NTPase will most likely work for SARS-CoV-2 as well. Specific examples of such inhibitors are shown in Table 9, and a more complete list is given in Table S5. Some trioxaadamantanetriol compounds, such as bananin and vanillinbananin, inhibited replication of SARS-CoV in cultured cells with low cytotoxicity.110,111 In addition, the plant-derived flavonoids myricetin and scutellarein, which are both found in tea, have been shown as active inhibitors with low toxicity.112,113 Unlike the above mentioned inhibitors, SSYA10–001 demonstrated helicase inhibition without affecting the cellular ATPase activity.114
Table 9. Small-Molecule Inhibitors of Helicase/NTPase80,103,110,113,115,116.
“*”: multiple activity measure values for one substance are from multiple references.
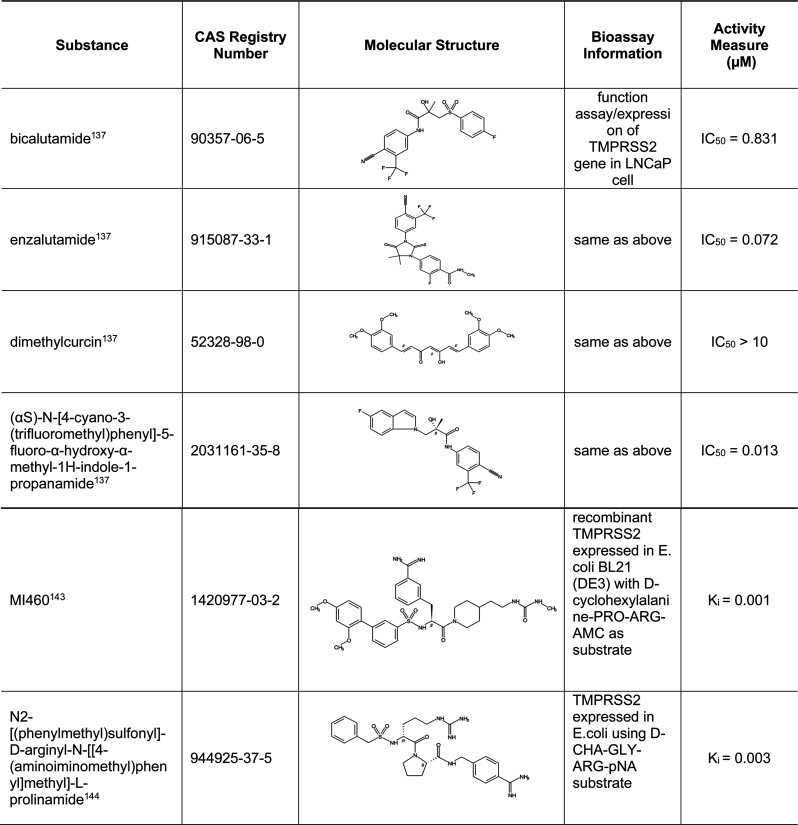
5.7. Small-Molecule Inhibitors of Human Protease TMPRSS2
Human serine protease TMPRSS2 is involved in S protein priming needed for the S2 segment of the S protein to mediate fusion of the viral envelope with the host cell membrane.7 Selected inhibitors are shown in Table 10, and a more complete list is given in Table S6. In addition to their inhibitory effect on TMPRSS2, these selected inhibitors are known to have other functions that may be beneficial in treating COVID-19. For example, bicalutamide, enzalutamide, dimethylcurcin, and CAS RN 2031161–35–8 are nonsteroidal antiandrogen drugs that were shown to inhibit TMPRSS2 expression.117 Since TMPRSS2 is an androgen-regulated gene that is overexpressed in prostate cancer,118 speculation has arisen that higher androgen levels could be the reason for more severe outcomes in men with COVID-19.119 In addition, inhibitors of androgen signaling have been shown to reduce ACE2 levels; therefore, these inhibitors may have dual functions affecting both ACE2 and TMPRSS2.120 Finally, compounds MI460 and CAS RN 944925–37–5 may also inhibit proinflammatory cytokines and block blood coagulation-related factors, respectively.121,122
Table 10. Small-Molecule Inhibitors of Human Protease TMPRSS2117,123,124.
5.8. Small-Molecule and Peptide Inhibitors of Human Protease Furin
The human protease furin is a ubiquitously expressed subtilisin/kexin-like proprotein convertase (PC) that cleaves the multibasic motif (RX(K/R)R↓) and activates/inactivates a variety of proteins including hormones, cytokines, and enzymes.125 Similar to TMPRSS2, furin is involved in priming viral S protein to mediate viral fusion with the host cell membrane and subsequent viral entry. Its cleavage site (RRAR↓) at the S1/S2 boundary of the SARS-CoV-2 and MERS-CoV S protein matches the minimal requirement of furin substrate sequence.126,127 Many furin inhibitors have been reported in the literature. Selected substances are shown in Table 11, and a more complete list is given in Table S7. The vast majority of furin inhibitors are peptides or peptidomimetics containing polyarginine or their derived analogs that bind to the catalytic site of furin.128 For instance, phenylacetyl-Arg-Val-Arg-4-amidinobenzylamide is one such substrate analog furin inhibitor. Further modification of this compound with additions of tert-leucine and a basic group, as represented by CAS RN 1788032–54–1,125 improved the potency of furin inhibition. Other examples are peptide inhibitors and peptidomimetics that were synthesized based on the RARRRKKRT inhibitory scaffold128 and the potent furin inhibitor Dec-RVKR-CMK that was shown to inhibit cleavage and viral replication in Vero cells.129
There are also nonpeptidic furin inhibitors, such as the guanidinylated aryl 2,5-dideoxystreptamine-derived compound represented by CAS RN 922732–52–3. This substance inhibited not only furin but also other PC family members (PC6B, PACE4, and PC7) without significant cytotoxicity to cells.130 Another inhibitor, oroxylin A, an O-methylated flavone natural product extracted from Scutellaria roots, has anti-inflammatory and anticoagulation activities, which may also be beneficial in treating COVID-19.131 Baicalein, a flavone from the roots of Scutellaria baicalensis, has been shown to inhibit Dengue virus replication in Vero cells and has also been reported recently to inhibit SARS-CoV-2 3CLpro.132 In addition, baicalein has antibacterial and anti-inflammatory activities.79 Although these inhibitors may be used to treat COVID-19 and its associated complications, more study is needed to ensure the safety of these compounds.
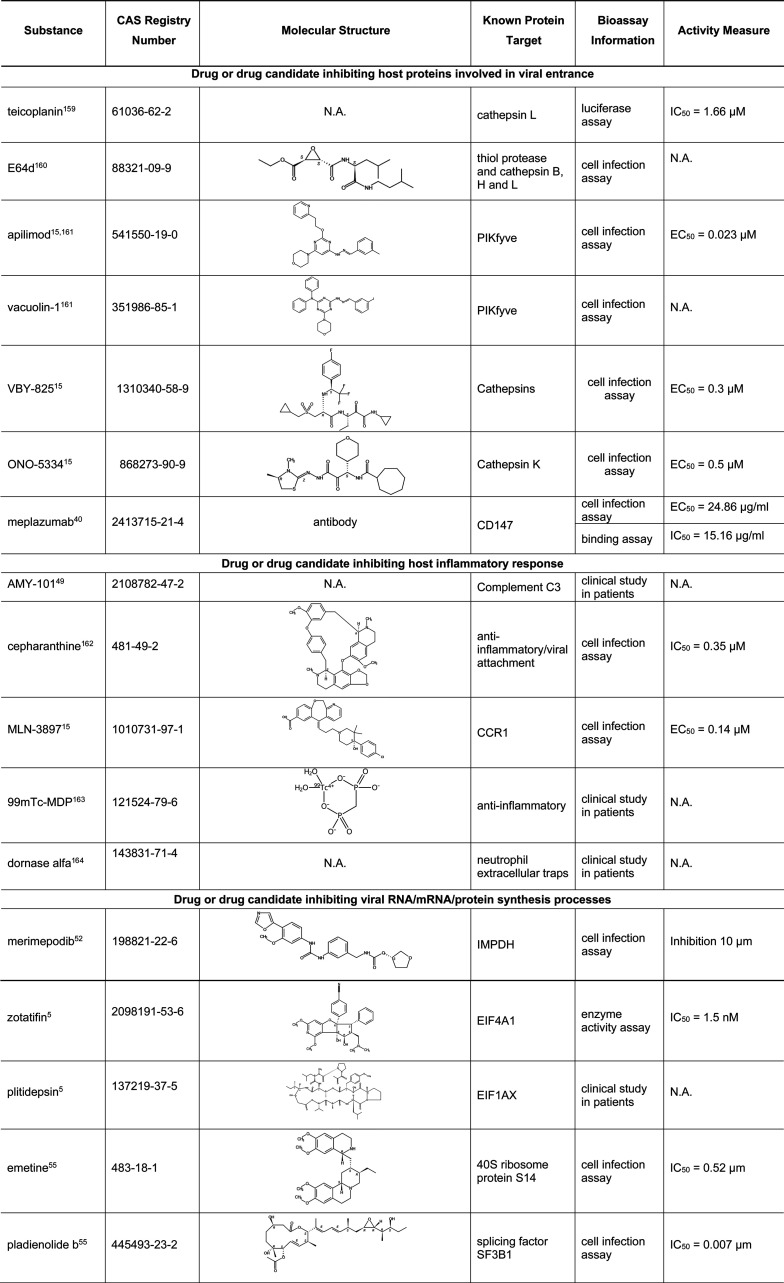
5.9. Small Molecules and Biologics Targeting Other Human Proteins Involved in SARS-CoV-2 Infection
In addition to TMPRSS2 and furin, there are many other human proteins as listed in Table 4 which have been shown to be involved in COVID-19. Table 12 lists several small molecules or biologics targeting these human proteins involved in different steps of SARS-CoV-2 infection. A number of these, including meplazumab, merimepodib, plitidepsin, niclosamide, dornase alfa, and AMY-101 are currently in COVID-19 clinical trials.
Table 12. Small Molecules and Biologics Targeting Other Human Proteins Involved in SARS-CoV-2 Infection5,15,40,49,52,55,139−144.
6. Summary and Perspectives
In light of the enormous amount of published information and rapidly evolving knowledge about COVID-19, this report systematically assembles and curates a large amount of data into one resource to support the ongoing research and development of COVID-19 therapeutics. Highlighted are notable journal articles and patents related to COVID-19, important viral and human protein targets, a high-level view of target–substance relationship in documents related to COVID-19, SARS, and MERS, as well as rich lists of target-based potential drug candidates for COVID-19 and related coronavirus infections. The potential drug candidates include both small- and large-molecule biologics. The small molecules are comprised of a wide variety of organic compounds, nucleotide analogs, and peptides, while the biologics are mainly antibodies along with a few recombinant proteins. More importantly, we report bioassay data with detailed structure–activity relationship information extracted from published studies. We hope this report will be valuable to the ongoing drug repurposing efforts and the discovery of new therapeutics with the potential for treating COVID-19. It is worth mentioning that although these preclinical studies provide important information the utility of the listed substances as drugs for COVID-19 or related coronavirus infections would ultimately rely on successful clinical trials.
In addition to the various wet-laboratory-based approaches, computational drug repurposing for COVID-19 also plays a significant role in accelerating therapeutic development for this and other diseases. In this approach, a variety of computational and clinical data are often used and analyzed together for drug repurposing.145 This approach can help to overcome the challenge of translating basic scientific findings to human applications, because these drugs have passed clinical safety and bioavailability testing, thereby increasing their chances for final approval.146 For example, in a high-throughput docking approach, after screening a chemical library built from FDA-approved drugs and compounds undergoing clinical trials, Cavasotto and Di Filippo identified several structurally diverse compounds that each displayed antiviral activity against SARS-CoV-2.147 Another structure-based virtual study suggested that toremifene, an FDA-approved estrogen receptor modulator for treating advanced breast cancer, may inhibit the SARS-CoV-2 S protein and methyltransferase/NSP14.148 Moreover, melatonin was identified by the network medicine approach as showing a significant association with reduced likelihood of SARS-CoV-2-positive test results.149 In addition, Zeng et al. demonstrated that deep learning is a powerful methodology to prioritize existing drugs for further investigation.150 Using a library of commercially available compounds, Elmezayen et al. discovered several potential inhibitors against 3CLpro or TMPRSS2 with virtual screening and further evaluated their absorption, distribution, metabolism, excretion, and toxicity (ADMET) profiles.151 It can be expected that computer-screening of compounds and modeling will be increasingly used in the discovery of drugs for COVID-19 and other viral infections to expedite the drug development process and lower its cost. Nevertheless, experimental evaluation of drug candidates’ efficacy in cell-based assays and animal model studies is still needed to confirm the suggested drug effects of these virtually selected molecules.
Although this paper focuses on individual therapeutics, therapy regimes that combine various drugs to target multiple pathological processes and/or molecular targets have been evaluated and may play an important role in treating COVID-19. Over 600 documents covered drug combination approaches for COVID-19 in the CAS scientific literature collection. These include studies on the well-publicized hydroxychloroquine and azithromycin combination and on remdesivir with numerous other drugs. Of the latter, one interesting paper that combined in silico and in vitro methods highlighted the combination of remdesivir with nitazoxanide.152 Since COVID-19 is often characterized by exaggerated inflammatory responses, anti-inflammatory treatments are often combined with antiviral agents. For example, Zhou et al. found that the mercaptopurine/melatonin and toremifene/emodin combinations were potentially of value in a computational network pharmacology study.153
Since COVID-19 patients with an underlying condition, such as cardiovascular disease or diabetes, are more likely to be hospitalized and have life-threatening conditions, it is very likely that COVID-19 patients receiving antiviral drugs are simultaneously on other medications for their pre-existing conditions. Therefore, it is also crucial that COVID-19 drugs given should be compatible with those medications that the patient is already taking in order to prevent undesirable drug–drug interactions.
Currently, it is unknown how long the COVID-19 crisis will last. As different parts of the world become increasingly interconnected, it seems likely that there will be additional pandemics in the years to come, and many of these will be of viral origin. We hope the current focus on antiviral agent research will lead to major breakthroughs and help us to be better prepared for future outbreaks.
Acknowledgments
We sincerely appreciate Jeffrey Smoot, Mark Johnson, Lihua Nie, David Leybman, Ilya Utkin, ChunAn Chen, and Philip Trinity for their technical assistance during preparation of this paper. We thank Susan Jervey for her careful editing. We are also very grateful to Manuel Guzman, Gilles George, Dana Albaiu, and Jeffrey Wilson for their encouragement and support.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsptsci.0c00074.
Table S1: a more complete list of human protein targets involved in COVID-19, Table S2: inhibitors of 3CLpro in SARS-CoV and MERS-CoV, Table S3: inhibitors of PLpro in SARS-CoV, Table S4: inhibitors of RdRp in positive-stranded ssRNA viruses, Table S5: inhibitors of helicase/NTPase in SARS-CoV, Table S6: inhibitors of human proteaseTMPRSS2, Table S7: inhibitors of human protease furin (XLSX)
Author Contributions
† Q.A.Z. and J.K.-W. contributed equally to this paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Zhu N.; Zhang D.; Wang W.; Li X.; Yang B.; Song J.; Zhao X.; Huang B.; Shi W.; Lu R.; Niu P.; Zhan F.; Ma X.; Wang D.; Xu W.; Wu G.; Gao G. F.; Tan W. (2020) A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus Disease (COVD-19) Dashboard, covid19.who.int (accessed on 6/25/2020).
- Lu R.; Zhao X.; Li J.; Niu P.; Yang B.; Wu H.; Wang W.; Song H.; Huang B.; Zhu N.; Bi Y.; Ma X.; Zhan F.; Wang L.; Hu T.; Zhou H.; Hu Z.; Zhou W.; Zhao L.; Chen J.; Meng Y.; Wang J.; Lin Y.; Yuan J.; Xie Z.; Ma J.; Liu W. J.; Wang D.; Xu W.; Holmes E. C.; Gao G. F.; Wu G.; Chen W.; Shi W.; Tan W. (2020) Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–74. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.; Siddique R.; Shereen M. A.; Ali A.; Liu J.; Bai Q.; Bashir N.; Xue M. (2020) Emergence of a Novel Coronavirus, Severe Acute Respiratory Coronavirus 2: Biology and Therapeutic Options. J. Clin. Microbiol. 58 (5), e00187-20 10.1128/JCM.00187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. E.; Jang G. M.; Bouhaddou M.; Xu J.; Obernier K.; White K. M.; O’Meara M. J.; Rezelj V. V.; Guo J. Z.; Swaney D. L.; et al. (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Ye G.; Shi K.; Wan Y.; Luo C.; Aihara H.; Geng Q.; Auerbach A.; Li F. (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581, 221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N. H.; Nitsche A.; Müller M. A.; Drosten C.; Pohlmann S. (2020) SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 is Blocked by a Clinically Proven Protease Inhibitor. Cell 181 (2), 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.; De Clercq E. (2020) Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discovery 19, 149–150. 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Morse J. S.; Lalonde T.; Xu S.; Liu W. R. (2020) Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV. ChemBioChem 21 (5), 730–738. 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y. M.; St. John S. E.; Mesecar A. D. (2015) The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antiviral Res. 115, 21–38. 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N., Yang X., Ye L., Chen K., Chan E. W.-C., Yang M., and Chen S. (2020) Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China. bioRxiv, DOI: 10.1101/2020.01.20.913368.
- Tay M. Z.; Poh C. M.; Rénia L.; MacAry P. A.; Ng L. F. P. (2020) The Trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374. 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. B.; June C. H. (2020) Cytokine release syndrome in severe COVID-19. Science (Washington, DC, U. S.) 368 (6490), 473–474. 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- Bikdeli B.; Madhavan M. V.; Jimenez D.; Chuich T.; Dreyfus I.; Driggin E.; Der Nigoghossian C.; Ageno W.; Madjid M.; Guo Y.; et al. (2020) COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-Up. J. Am. Coll. Cardiol. 75 (23), 2950–2973. 10.1016/j.jacc.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva L.; Yuan S.; Yin X.; Martin-Sancho L.; Matsunaga N.; Pache L.; Burgstaller-Muehlbacher S.; De Jesus P. P.; Teriete P.; Hull M. V.; et al. (2020) Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Cao R.; Zhang H.; Liu J.; Xu M.; Hu H.; Li Y.; Zhao L.; Li W.; Sun X.; Yang X.; Shi Z.; Deng F.; Hu Z.; Zhong W.; Wang M. (2020) The anti-influenza virus drug, Arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discovery 6, 28. 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M.; Matsuyama S.; Li X.; Takeda M.; Kawaguchi Y.; Inoue J.-I.; Matsuda Z. (2016) Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob. Agents Chemother. 60 (11), 6532–6539. 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trial of Inhaled anti-viral (SNG001) for SARS-CoV-2 (COVID-19) Infection. Clinical Trial no. NCT04385095, https://clinicaltrials.gov/ct2/show/NCT04385095 (accessed on 6/17/2020).
- Efficacy and Safety of Novel Treatment Options for Adults With COVID-19 Pneumonia. Clinical Trial no. NCT04345289, https://clinicaltrials.gov/ct2/show/NCT04345289 (accessed on 6/17/2020).
- Effect of Treatments in Patients Hospitalized for Severe COVID-19 Pneumonia: A Multicenter Cohort Study. Clinical Trial no. NCT04365764, https://clinicaltrials.gov/ct2/show/NCT04365764 (accessed on 6/17/2020).
- Roschewski M.; Lionakis M. S.; Sharman J. P.; Roswarski J.; Goy A.; Monticelli M. A.; Roshon M.; Wrzesinski S. H.; Desai J. V.; Zarakas M. A.; Collen J.; Rose K.; Hamdy A.; Izumi R.; Wright G. W.; Chung K. K.; Baselga J.; Staudt L. M.; Wilson W. H. (2020) Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci. Immunol. 5 (48), eabd0110 10.1126/sciimmunol.abd0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W. S., Emberson J. R., Mafham M., Bell J. L., Linsell L., Staplin N., Brightling C., Ustianowski A., and Elmahi E.. et al. Dexamethasone in Hospitalized Patients with Covid-19 - Preliminary Report. N. Engl. J. Med., 2020, 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randomised Evaluation of COVID-19 Therapy (RECOVERY). Clinical Trial no. NCT04381936. https://clinicaltrials.gov/ct2/show/NCT04381936 (accessed on 7/18/2020).
- U.S. Food & Drug Administration . (2020) Fact Sheet for Health Care Providers Emergency Use Authorization (EUA) of Remdesivir (GS-5734). https://www.fda.gov/media/137566/download.
- Glenmark Initiates Phase 3 Clinical Trials on Antiviral Favipiravir for COVID-10 Patients in India. Glenmark Pharmaceuticals Ltd., News release, May 12, 2020. www.prnewswire.com/in/news-releases/glenmark-initiates-phase-3-clinical-trials-on-antiviral-favipiravir-for-covid-19-patients-in-india-800133083.html. [Google Scholar]
- Russian Ministry of Health approves the first COVID-19 drug Avifavir produced by JV of RDIF and ChemRar. Russian Direct Investment Fund News release, May 30, 2020. rdif.ru/Eng_fullNews/5220/. [Google Scholar]
- Geleris J.; Sun Y.; Platt J.; Zucker J.; Baldwin M.; Hripcsak G.; Labella A.; Manson D.; Kubin C.; Barr G.; Sobieszczyk M.; Schluger N. (2020) Observational Study of Hydroxychloroquine in Hospitalized Patients with Covid-19. N. Engl. J. Med. 382, 2411–2418. 10.1056/NEJMoa2012410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitja O.; Corbacho-Monne M.; Ubals M.; Tebe C.; Penafiel J.; Tobias A.; Ballana E.; Alemany A.; Riera-Marti N.; Perez C.; et al. (2020) Hydroxychloroquine for Early Treatment of Adults with Mild Covid-19: A Randomized Controlled Trial. Clin. Infect. Dis. ciaa1009. 10.1093/cid/ciaa1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashour Z. T., Riaz M., Garbati M., Aldosary O., Tlayjeh H., Gerberi D., Murad M. H., Sohail M. R., Kashour T., and Tleyjeh I. M. (2020) Efficacy of Chloroquine or Hydroxychloroquine in COVID-19 Patients: A Systematic Review and Meta-Analysis. medRxiv 2020.07.12.20150110, DOI: 10.1101/2020.07.12.20150110. [DOI] [PMC free article] [PubMed]
- U.S. Food & Drug Administration . (2020) Revoke of EUA for emergency use of oral formulations of chloroquine phosphate (CQ) and hydroxychloroquine sulfate (HCQ). https://www.fda.gov/media/138945/download.
- U.S. Food & Drug Administration . (2020) FDA cautions against use of hydroxychloroquine or chloroquine for COVID-19 outside of the hospital setting or a clinical trial due to risk of heart rhythm problems. https://www.fda.gov/drugs/drug-safety-and-availability/fda-cautions-against-use-hydroxychloroquine-or-chloroquine-covid-19-outside-hospital-setting-or (accessed on 7/18/2020).
- Liu C.; Zhou Q.; Li Y.; Garner L. V.; Watkins S. P.; Carter L. J.; Smoot J.; Gregg A. C.; Daniels A. D.; Jervey S.; Albaiu D. (2020) Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent. Sci. 6 (3), 315–331. 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R.; Perlman S. (2015) Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 1282, 1–23. 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E. J.; van der Meer Y.; Zevenhoven-Dobbe J.; Onderwater J. J. M.; van der Meulen J.; Koerten H. K.; Mommaas A. M. (2006) Ultrastructure and Origin of Membrane Vesicles Associated with the Severe Acute Respiratory Syndrome Coronavirus Replication Complex. J. Virol. 80 (12), 5927–5940. 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M.; Imbert I.; Subissi L.; Gluais L.; Canard B.; Decroly E. (2012) RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U. S. A. 109 (24), 9372–9377. 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M.; Huang C.; Shirato K.; Makino S.; Taguchi F. (2016) The Contribution of the Cytoplasmic Retrieval Signal of Severe Acute Respiratory Syndrome Coronavirus to Intracellular Accumulation of S Proteins and Incorporation of S Protein Into Virus-Like Particles. J. Gen. Virol. 97 (8), 1853–1864. 10.1099/jgv.0.000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt A. L.; Larson B. J.; Hogue B. G. A. (2010) Conserved Domain in the Coronavirus Membrane Protein Tail Is Important for Virus Assembly. J. Virol. 84 (21), 11418–11428. 10.1128/JVI.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M.; Yount B.; Heise M.; Kopecky-Bromberg S. A.; Palese P.; Baric R. S. (2007) SARS-CoV ORF6 Antagonizes STAT1 Function by Sequestering Nuclear Import Factors on the rER/Golgi membrane. J. Virol. 81 (18), 9812–9824. 10.1128/JVI.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H.; Pillat M. (2020) CD147 as a Target for COVID-19 Treatment: Suggested Effects of Azithromycin and Stem cell engagement. Stem Cell Rev. Rep. 16 (3), 434–440. 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhou Y., Lian J., Zhang Z., Du P., Gong L., and Zhang Y., et al. (2020) SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv, DOI: 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed]
- Ou X.; Liu Y.; Lei X.; Li P.; Mi D.; Ren L.; Guo L.; Guo R.; Chen T.; Hu J.; Xiang Z.; Mu Z.; Chen X.; Chen J.; Hu K.; Jin Q.; Wang J.; Qian Z. (2020) Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11 (1), 1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozzini C.; Girelli D. (2020) The role of Neutrophil Extracellular Traps in Covid-19: Only an Hypothesis or a Potential new field of research?. Thromb. Res. 191, 26–27. 10.1016/j.thromres.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker H., and Parkkila S. (2020) Bioinformatic characterization of angiotensin-converting enzyme 2, the entry receptor for SARS-CoV-2. bioRxiv, DOI: 10.1101/2020.04.13.038752. [DOI] [PMC free article] [PubMed]
- Wu A., Niu P., Wang L., Zhou H., Zhao X., Wang W., Wang J., Ji C., and Ding X.. et al. (2020) Mutations, recombination and insertion in the evolution of 2019-nCoV. bioRxiv, DOI: 10.1101/2020.02.29.971101.
- Shang J.; Wan Y.; Luo C.; Ye G.; Geng Q.; Auerbach A.; Li F. (2020) Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 117 (21), 11727–11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle K. D., Minter R., Waugh K. A., Araya P., Ludwig M., Sempeck C., Smith K., Andrysik Z., Burchill M. A., and Tamburini B. A. J.. et al. (2020) JAK1 inhibition blocks lethal sterile immune responses: implications for COVID-19 therapy. bioRxiv, DOI: 10.1101/2020.04.07.024455. [DOI] [PMC free article] [PubMed]
- Cantini F.; Niccoli L.; Matarrese D.; Nicastri E.; Stobbione P.; Goletti D. (2020) Baricitinib therapy in COVID-19: A pilot study on safety and clinical impact. J. Infect. 81, 318. 10.1016/j.jinf.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perricone C.; Triggianese P.; Bartoloni E.; Cafaro G.; Bonifacio A. F.; Bursi R.; Perricone R.; Gerli R. (2020) The anti-viral facet of anti-rheumatic drugs: Lessons from COVID-19. J. Autoimmun. 111, 102468. 10.1016/j.jaut.2020.102468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastaglio S.; Ruggeri A.; Risitano A. M.; Angelillo P.; Yancopoulou D.; Mastellos D. C.; Huber-Lang M.; Piemontese S.; Assanelli A.; Garlanda C.; Lambris J. D.; Ciceri F. (2020) The first case of COVID-19 treated with the complement C3 inhibitor AMY-101. Clin. Immunol. (Amsterdam, Neth.) 215, 108450. 10.1016/j.clim.2020.108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Shen C., Li J., Yuan J., Yang M., Wang F., Li G., Li Y., Xing L., and Peng L.. et al. (2020) Exuberant elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 2 infection is associated with disease severity and fatal outcome. medRxiv, DOI: 10.1101/2020.03.02.20029975.
- Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J. A., Blair C., Weber A., Barnes B. J., and Egeblad M.. et al. (2020) Neutrophil extracellular traps (NETs) as markers of disease severity in COVID-19. medRxiv, DOI: 10.1101/2020.04.09.20059626. [DOI] [PMC free article] [PubMed]
- Bukreyeva N., Mantlo E. K., Sattler R. A., Huang C., Paessler S., and Zeldis J. (2020) The IMPDH inhibitor merimepodib suppresses SARS-CoV-2 replication in vitro. bioRxiv, DOI: 10.1101/2020.04.07.028589.
- Müller C.; Schulte F. W.; Lange-Grünweller K.; Obermann W.; Madhugiri R.; Pleschka S.; Ziebuhr J.; Hartmann R. K.; Grünweller A. (2018) Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antiviral Res. 150, 123–129. 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K.; Swett-Tapia C.; van den Worm S. H. E.; te Velthuis A. J. W.; Koster A. J.; Mommaas A. M.; Snijder E. J.; Kikkert M. (2010) Integrity of the Early Secretory Pathway promotes, but Is Not Required for, Severe Acute Respiratory Syndrome Coronavirus RNA Synthesis and Virus-Induced Remodeling of Endoplasmic Reticulum Membranes. J. Virol. 84 (2), 833–846. 10.1128/JVI.01826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D.; Klann K.; Koch B.; Widera M.; Krause D.; Ciesek S.; Cinatl J.; Muench C. (2020) Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 583, 469. 10.1038/s41586-020-2332-7. [DOI] [PubMed] [Google Scholar]
- Srinivasan S.; Cui H.; Gao Z.; Liu M.; Lu S.; Mkandawire W.; Narykov O.; Sun M.; Korkin D. (2020) Structural Genomics of SARS-CoV-2 Indicates Evolutionary Conserved Functional Regions of Viral Proteins. Viruses 12 (4), 360. 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Hurst B., Hu Y., Szeto T., Tarbet B., and Wang J. (2020) Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. bioRxiv, DOI: 10.1101/2020.04.20.051581. [DOI] [PMC free article] [PubMed]
- Vuong W., Khan M. B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H. A., McKay R. T., van Belkum M. J., and Joyce M.. et al. (2020) Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. bioRxiv, DOI: 10.1101/2020.05.03.073080. [DOI] [PMC free article] [PubMed]
- Zhang L.; Lin D.; Sun X.; Curth U.; Drosten C.; Sauerhering L.; Becker S.; Rox K.; Hilgenfeld R. (2020) Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (Washington, DC, U. S.) 368 (6489), 409–412. 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W.; Zhang B.; Jiang X.-M.; Su H.; Li J.; Zhao Y.; Xie X.; Jin Z.; Peng J.; Liu F.; Li C.; Li Y.; Bai F.; Wang H.; Cheng X.; Cen X.; Hu S.; Yang X.; Wang J.; Liu X.; Xiao G.; Jiang H.; Rao Z.; Zhang L.-K.; Xu Y.; Yang H.; Liu H. (2020) Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science (Washington, DC, U.S.) 368 (6497), 1331–1335. 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Li X., Huang Y.-Y., Wu Y., Zhou L., Liu R., Wu D., Zhang L., Liu H., and Xu X.. et al. (2020) Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. bioRxiv, DOI: 10.1101/2020.03.23.004580. [DOI] [PMC free article] [PubMed]
- Liu X., Li Z., Liu S., Chen Z., Sun J., Zhao Z., Huang Y.-Y., Zhang Q., Wang J., and Shi Y.. et al. (2020) Therapeutic effects of dipyridamole on COVID-19 patients with coagulation dysfunction. medRxiv, DOI: 10.1101/2020.02.27.20027557.
- Fintelman-Rodrigues N., Sacramento C. Q., Lima C. R., da Silva F. S., Ferreira A. C., Mattos M., de Freitas C. S., Soares V. C., da Silva Gomes Dias S., Temerozo J. R., Miranda M., Matos A. R., Bozza F. A., Carels N., Alves C. R., Siqueira M. M., Bozza P. T., and Souza T. M. L. (2020) Atazanavir inhibits SARS-CoV-2 replication and pro-inflammatory cytokine production. bioRxiv, DOI: 10.1101/2020.04.04.020925.
- Yamamoto N., Matsuyama S., Hoshino T., and Yamamoto N. (2020) Nelfinavir inhibits replication of severe acute respiratory syndrome coronavirus 2 in vitro. bioRxiv, DOI: 10.1101/2020.04.06.026476.
- Chen H., Zhang Z., Wang L., Huang Z., Gong F., Li X., Chen Y., and Wu J. J. (2020) First clinical study using HCV protease inhibitor to treat naive and experienced COVID-19 patients. medRxiv, DOI: 10.1101/2020.03.22.20034041. [DOI] [PMC free article] [PubMed]
- Sargsyan K., Lin C.-C., Chen T., Grauffel C., Chen Y.-P., Yang W.-Z., Yuan H. S., and Lim C. (2020) Multi-Targeting of Functional Cysteines in Multiple Conserved SARS-CoV-2 Domains by Clinically Safe Zn-ejectors. ChemRxiv, DOI: 10.26434/chemrxiv.12179037. [DOI] [PMC free article] [PubMed]
- Jin Z.; Du X.; Xu Y.; Deng Y.; Liu M.; Zhao Y.; Zhang B.; Li X.; Zhang L.; Peng C.; Duan Y.; Yu J.; Wang L.; Yang K.; Liu F.; Jiang R.; Yang X.; You T.; Liu X.; Yang X.; Bai F.; Liu H.; Liu X.; Guddat L. W.; Xu W.; Xiao G.; Qin C.; Shi Z.; Jiang H.; Rao Z.; Yang H. (2020) Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature 582, 289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jin Z.; Zhao Y.; Sun Y.; Zhang B.; Wang H.; Wu Y.; Zhu Y.; Zhu C.; Hu T.; Du X.; Duan Y.; Yu J.; Yang X.; Yang X.; Yang K.; Liu X.; Guddat L. W.; Xiao G.; Zhang L.; Yang H.; Rao Z. (2020) Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 27, 529–532. 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- Wen C.-C; Kuo Y.-H.; Jan J.-T.; Liang P.-H.; Wang S.-Y.; Liu H.-G.; Lee C.-K.; Chang S.-T.; Kuo C.-J.; Lee S.-S.; Hou C.-C.; Hsiao P.-W.; Chien S.-C.; Shyur L.-F.; Yang N.-S. (2007) Specific Plant Terpenoids and Lignoids Possess Potent Antiviral Activities against Severe Acute Respiratory Syndrome Coronavirus. J. Med. Chem. 50 (17), 4087–4095. 10.1021/jm070295s. [DOI] [PubMed] [Google Scholar]
- Liu X.; Jutooru I.; Lei P.; Kim K. H.; Lee S.; Brents L. K.; Prather P. L.; Safe S. (2012) Betulinic acid targets YY1 and ErbB2 through cannabinoid receptor-dependent disruption of microRNA-27a:ZBTB10 in breast cancer. Mol. Cancer Ther. 11 (7), 1421–1431. 10.1158/1535-7163.MCT-12-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C.; Sadek B.; Goyal S. N.; Sinha S.; Kamal M. A.; Ojha S. (2015) Small Molecules from Nature Targeting G-Protein Coupled Cannabinoid Receptors: Potential Leads for Drug Discovery and Development. Evidence-Based Complementary Altern. Med. 2015, 238482 10.1155/2015/238482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. Y.; Park J.; Kim P. S.; Yoo E. S.; Baik K. U.; Park M. H. (2001) Savinin, a lignan from Pterocarpus santalinus inhibits tumor necrosis factor- SSproduction and T cell proliferation. Biol. Pharm. Bull. 24 (2), 167–171. 10.1248/bpb.24.167. [DOI] [PubMed] [Google Scholar]
- Rossi F.; Tortora C.; Argenziano M.; Di Paola A.; Punzo F. (2020) Cannabinoid Receptor Type 2: A Possible Target in SARS-CoV-2 (CoV-19) Infection?. Int. J. Mol. Sci. 21 (11), 3809. 10.3390/ijms21113809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkatti P.; Pandey R.; Rieder S. A.; Hegde V. L.; Nagarkatti M. (2009) Cannabinoids as novel anti-inflammatory drugs. Future Med. Chem. 1 (7), 1333–1349. 10.4155/fmc.09.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann M.; Maini R. N.; Woody J. N.; Holgate S. T.; Winter G.; Rowland M.; Richards D.; Hussell T. (2020) Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet 395 (10234), 1407–1409. 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirois S.; Zhang R.; Gao W.; Gao H.; Li Y.; Zheng H.; Wei D.-Q. (2007) Discovery of Potent Anti-SARS-CoV MPro Inhibitors. Curr. Comput.-Aided Drug Des. 3 (3), 191–200. 10.2174/157340907781695440. [DOI] [Google Scholar]
- Du Q. S.; Wang S. Q.; Zhu Y.; Wei D.-Q.; Guo H.; Sirois S.; Chou K.-C. (2004) Polyprotein cleavage mechanism of SARS CoV Mpro and chemical modification of the octapeptide. Peptides 25 (11), 1857–1864. 10.1016/j.peptides.2004.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Ye F., Sun Q., Liang H., Li C., Lu R., Huang B., Tan W., and Lai L. (2020) Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. bioRxiv, DOI: 10.1101/2020.04.10.035824. [DOI] [PMC free article] [PubMed]
- Su H., Yao S., Zhao W., Li M., Liu J., Shang W., Xie H., Ke C., Gao M., and Yu K.. et al. (2020) Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. bioRxiv, DOI: 10.1101/2020.04.13.038687.
- Wu Y. S.; Lin W.-H.; Hsu J. T.-A.; Hsieh H.-P. (2006) Antiviral drug discovery against SARS-CoV. Curr. Med. Chem. 13 (17), 2003–2020. 10.2174/092986706777584988. [DOI] [PubMed] [Google Scholar]
- World Health Organization Model List of Essential Medicines, 21st list, 2019, https://apps.who.int/iris/bitstream/handle/10665/325771/WHO-MVP-EMP-IAU-2019.06-eng.pdf (accessed on 6/7/2020).
- Chou C.-Y.; Chien C.-H.; Han Y.-S.; Prebanda M. T.; Hsieh H.-P.; Turk B.; Chang G.-G.; Chen X. (2008) Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem. Pharmacol. (Amsterdam, Neth.) 75 (8), 1601–1609. 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K.; Takayama J.; Aubin Y.; Ratia K.; Chaudhuri R.; Baez Y.; Sleeman K.; Coughlin M.; Nichols D. B.; Mulhearn D. C.; Prabhakar B. S.; Baker S. C.; Johnson M. E.; Mesecar A. D. (2009) Structure-Based Design, Synthesis, and Biological Evaluation of a Series of Novel and Reversible Inhibitors for the Severe Acute Respiratory Syndrome-Coronavirus Papain-Like Protease. J. Med. Chem. 52 (16), 5228–5240. 10.1021/jm900611t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K.; Takayama J.; Rao K. V.; Ratia K.; Chaudhuri R.; Mulhearn D. C.; Lee H.; Nichols D. B.; Baliji S.; Baker S. C.; Johnson M. E.; Mesecar A. D. (2010) Severe Acute Respiratory Syndrome Coronavirus Papain-like Novel Protease Inhibitors: Design, Synthesis, Protein-Ligand X-ray Structure and Biological Evaluation. J. Med. Chem. 53 (13), 4968–4979. 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratia K.; Pegan S.; Takayama J.; Sleeman K.; Coughlin M.; Baliji S.; Chaudhuri R.; Fu W.; Prabhakar B. S.; Johnson M. E.; et al. (2008) A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 105 (42), 16119–16124. 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. K., and Takayama J. (2010) Compounds and methods for treating respiratory diseases, WO2010022355A1, World Intellectual Property Organization.
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., and Zhang L.. et al. (2020) Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target. bioRxiv, DOI: 10.1101/2020.03.16.993386.
- Chien M., Anderson T. K., Jockusch S., Tao C., Kumar S., Li X., Russo J. J., Kirchdoerfer R. N., and Ju J. (2020) Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase. bioRxiv, DOI: 10.1101/2020.03.18.997585. [DOI] [PMC free article] [PubMed]
- Jockusch S., Tao C., Li X., Anderson T. K., Chien M., Kumar S., Russo J. J., Kirchdoerfer R. N., and Ju J. (2020) Triphosphates of the two components in DESCOVY and TRUVADA are inhibitors of the SARS-CoV-2 polymerase. bioRxiv, DOI: 10.1101/2020.04.03.022939.
- Jockusch S.; Tao C.; Li X.; Anderson T. K.; Chien M.; Kumar S.; Russo J. J.; Kirchdoerfer R. N.; Ju J. (2020) A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses causing SARS and COVID-19. Antiviral Res. 180, 104857. 10.1016/j.antiviral.2020.104857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P.; Sims A. C.; Zhou S.; Graham R. L.; Pruijssers A. J.; Agostini M. L.; Leist S. R.; Schafer A.; Dinnon K. H. III; Stevens L. J.; Chappell J. D.; Lu X.; Hughes T. M.; George A. S.; Hill C. S.; Montgomery S. A.; Brown A. J.; Bluemling G. R.; Natchus M. G.; Saindane M.; Kolykhalov A. A.; Painter G.; Harcourt J.; Tamin A.; Thornburg N. J.; Swanstrom R.; Denison M. R.; Baric R. S. (2020) An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 12 (541), eabb5883 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S.; Liu M.; Wang C.; Xu W.; Lan Q.; Feng S.; Qi F.; Bao L.; Du L.; Liu S.; Qin C.; Sun F.; Shi Z.; Zhu Y.; Jiang S.; Lu L. (2020) Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 30 (4), 343–355. 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.; Yu D.; Yan H.; Chong H.; He Y. (2020) Design of Potent Membrane Fusion Inhibitors Against SARS-CoV-2, an Emerging Coronavirus With High Fusogenic Activity. J. Virol. 94, e00635-20. 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Pomplun S., Loftis A. R., Loas A., and Pentelute B. L. (2020) Investigation of ACE2 N-terminal fragment binding to SARS-CoV-2 Spike RBD. bioRxiv, DOI: 10.1101/2020.03.19.999318.
- Li Y., Wang H., Tang X., Ma D., Du C., Wang Y., Pan H., Zou Q., Zheng J., and Xu L.. et al. (2020) Potential host range of multiple SARS-like coronaviruses and an improved ACE2-Fc variant that is potent against both SARS-CoV-2 and SARS-CoV-1. bioRxiv, DOI: 10.1101/2020.04.10.032342.
- Monteil V.; Kwon H.; Prado P.; Hagelkruys A.; Wimmer R. A.; Stahl M.; Leopoldi A.; Garreta E.; Hurtado del Pozo C.; Prosper F.; Romero J. P.; Wirnsberger G.; Zhang H.; Slutsky A. S.; Conder R.; Montserrat N.; Mirazimi A.; Penninger J. M. (2020) Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 181 (4), 905–913. 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Zhou P., Fan T., Wu Y., Zhang J., Shi X., Shang W., Fang L., Jiang X., and Shi J.. et al. (2020) Immunoglobulin fragment F(ab′)2 against RBD potently neutralizes SARS-CoV-2 in vitro. bioRxiv, DOI: 10.1101/2020.04.07.029884. [DOI] [PMC free article] [PubMed]
- Cao Y.; Su B.; Guo X.; Sun W.; Deng Y.; Bao L.; Zhu Q.; Zhang X.; Zheng Y.; Geng C.; Chai X.; He R.; Li X.; Lv Q.; Zhu H.; Deng W.; Xu Y.; Wang Y.; Qiao L.; Tan Y.; Song L.; Wang G.; Du X.; Gao N.; Liu J.; Xiao J.; Su X.; Du Z.; Feng Y.; Qin C.; Qin C.; Jin R.; Xie X. S. (2020) Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 182, 73. 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Liu X., Wang C., Zhang X., Ren L., Jin Q., Wang J., and Yang W. (2020) Humanized single domain antibodies neutralize SARS-CoV-2 by targeting spike receptor binding domain. bioRxiv, DOI: 10.1101/2020.04.14.042010. [DOI] [PMC free article] [PubMed]
- Wang C.; Li W.; Drabek D.; Okba N. M. A.; van Haperen R.; Osterhaus A. D. M. E.; van Kuppeveld F. J. M.; Haagmans B. L.; Grosveld F.; Bosch B.-J. (2020) A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 11 (1), 2251. 10.1038/s41467-020-16452-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Kral P. (2020) Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano 14, 5143–5147. 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbing J.; Phelan A.; Griffin I.; Tucker C.; Oechsle O.; Smith D.; Richardson P. (2020) COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 20, 400. 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. (2020) COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 20, 269. 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott T. R.; Dhamdhere G.; Liu Y.; Lin X.; Goudy L.; Zeng L.; Chemparathy A.; Chmura S.; Heaton N. S.; Debs R.; et al. (2020) Development of CRIPSR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell 181, 865. 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A.-T.; Gentile F.; Hsing M.; Ban F.; Cherkasov A. (2020) Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inf. 39, 2000028. 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., and Smith J. C. (2020) Repurposing therapeutics for COVID-19: Supercomputerbased docking to the SARSCoV-2 viral spike protein and viral spike protein-human ACE2 interface. ChemRxiv, DOI: 10.26434/chemrxiv.11871402.v4.
- Liu W., Liu W., Hu J., Lan H., and Liu W. (2020) Application of silver monoethyl fumarate in resisting novel coronavirus infection, CN111184708A, People's Republic of China: (in Chinese).
- Luo H., Huang Y., Li Z., and Zhou Q. (2020) Use of tolfenamic acid or a pharmaceutically acceptable salt thereof in the preparation of medicament for preventing and/or treating novel coronavirus inflammation, CN111184707A, People's Republic of China: (in Chinese).
- , Shi Y., Tan W., Wang M., Ye F., Peng Q., Huang B., Wang W., Su J., Zhao J., and Yuan B.. et al. (2020) Application of LTX-315 in preparing products for inhibiting coronavirus, CN111150833A, People's Republic of China: (in Chinese).
- Peng G., Liu Z., Chen X., and Tian D. (2020) Application of GS-441524 in preparing novel coronavirus SARS-CoV-2 inhibitor, CN111135184A, People's Republic of China: (in Chinese).
- Peng G., Liu Z., Chen X., and Tian D. (2020) Application of GC376 in preparation of novel coronavirus SARS-CoV-2 inhibitor, CN111135167A, People's Republic of China: (in Chinese).
- Peng G., Liu Z., Chen X., and Tian D. (2020) Pharmaceutical composition consisting of GC376 and GS-441524 and application thereof in inhibiting novel coronavirus, CN111135166A, People's Republic of China: (in Chinese).
- Wang G., Zhou J., Ye L., Sun Y., Jiang Z., Li X., Luo Q., and Xu L. (2020) Application of 2019-nCoV 3CL hydrolase inhibitor and IL-6 monoclonal antibody in preparing medicament or treating coronavirus disease 2019, CN111053909A, People's Republic of China: (in Chinese).
- Jin X., Chen P., and Cai Y. (2020) Composition containing benzylisoquinoline alkaloid and trans-resveratrol for treating coronavirus infection, CN110960532A, People's Republic of China: (in Chinese).
- Chen G., Wei Z., Zhao Y., Wang X., and Gu X. (2020) Overexpression ACE2 mesenchyma cell in the preparation of medicine for treating new coronavirus application of drugs and preparation method thereof, CN111166768A, People's Republic of China: (in Chinese).
- Weng B., and Li L. (2020) Preparation method of gene therapy product for treating COVID-19, CN111172195A, People's Republic of China: (in Chinese).
- Zhao F., Qin L., Wu J., Wang D., Dong B., Zhang F., and Wu X. (2020) A human SARS-CoV-2 monoclonal antibody and preparation method and application thereof, CN111153991A, People's Republic of China: (in Chinese).
- Hu R., Dong C., Zhang P., Zhang Z., Li Q., Wang X., Du Y., and Du H. (2020) SARS-CoV-2 Small-interfering nucleic acid, and its application for preparing pharmaceutical composition for preventing and/or treating new coronavirus pneumonia, CN111139242A, People's Republic of China: (in Chinese).
- Dong C., Hu R., Zhang P., Du Y., Wang X., Zhang Z., Li Q., and Du H. (2020) Small interfering nucleic acid for inhibiting new coronavirus and its composition and application, CN111139241A, People's Republic of China: (in Chinese).
- Kim S. C. (2020) COVID-19 virus customized triple knockout DNA treatment, KR2020032050, Korea: (in Korean).
- Li W., Drelich A., Martinez D. R., Gralinski L., Chen C., Sun Z., Schäfer A., Leist S. R., Liu X., and Zhelev D.. et al. (2020) Rapid selection of a human monoclonal antibody that potently neutralizes SARS-CoV-2 in two animal models. bioRxiv, DOI: 10.1101/2020.05.13.093088.
- Pinto D.; Park Y.; Beltramello M.; Walls A. C.; Tortorici M. A.; Bianchi S.; Jaconi S.; Culap K.; Zatta F.; De Marco A.; Peter A.; Guarino B.; Spreafico R.; Cameroni E.; Case J. B.; Chen R. E.; Havenar-Daughton C.; Snell G.; Telenti A.; Virgin H. W.; Lanzavecchia A.; Diamond M. S.; Fink K.; Veesler D.; Corti D. (2020) Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Kao R. Y.; Tsui W. H. W.; Lee T. S. W.; Tanner J. A.; Watt R. M.; Huang J.-D.; Hu L.; Chen G.; Chen Z.; Zhang L.; He T.; Chan K.-H.; Tse H.; To A. P. C.; Ng L. W. Y.; Wong B. C. W.; Tsoi H.-W.; Yang D.; Ho D. D.; Yuen K.-Y. (2004) Identification of Novel Small-Molecule Inhibitors of Severe Acute Respiratory Syndrome-Associated Coronavirus by Chemical Genetics. Chem. Biol. (Oxford, U. K.) 11 (9), 1293–1299. 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L.; Li Z.; Yuan K.; Qu X.; Chen J.; Wang G.; Zhang H.; Luo H.; Zhu L.; Jiang P.; Chen L.; Shen Y.; Luo M.; Zuo G.; Hu J.; Duan D.; Nie Y.; Shi X.; Wang W.; Han Y.; Li T.; Liu Y.; Ding M.; Deng H.; Xu X. (2004) Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol. 78 (20), 11334–11339. 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane J. S., Chatterjee A., Kumar A., and Ray S. (2020) Targeting SARS-CoV-2 Spike Protein of COVID-19 with Naturally Occurring Phytochemicals: An in Silco Study for Drug Development. ChemRxiv, DOI: 10.26434/chemrxiv.12094203.v1. [DOI] [PMC free article] [PubMed]
- Jo S.; Kim S.; Shin D. H.; Kim M.-S. (2020) Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 35 (1), 145. 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu I.-J.; Kao C.-L.; Hsieh S.-C.; Wey M.-T.; Kan L.-S.; Wang W.-K. (2009) Identification of a minimal peptide derived from heptad repeat (HR) 2 of spike protein of SARS-CoV and combination of HR1-derived peptides as fusion inhibitors. Antiviral Res. 81 (1), 82–87. 10.1016/j.antiviral.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T.-Y.; Wu S.-L.; Chen J.-C.; Wei Y.-C.; Cheng S.-E.; Chang Y.-H.; Liu H.-J.; Hsiang C.-Y. (2006) Design and biological activities of novel inhibitory peptides for SARS-CoV spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 69 (2), 70–76. 10.1016/j.antiviral.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto F. K. (2020) The Proteins of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS CoV 2 or n-COV19), the Cause of COVID 19. Protein J. 39, 198–216. 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesel A. J. (2012) An update on the bananins: anti-RNA-viral agents with unique structural signature. Anti-Infect. Agents 11 (1), 1–21. 10.2174/22113626130102. [DOI] [Google Scholar]
- Tanner J. A.; Zheng B.-J.; Zhou J.; Watt R. M.; Jiang J.-Q.; Wong K.-L.; Lin Y.-P.; Lu L.-Y.; He M.-L.; Kung H.-F.; Kesel A. J.; Huang J.-D. (2005) The Adamantane-Derived Bananins Are Potent Inhibitors of the Helicase Activities. Chem. Biol. (Oxford, U. K.) 12 (3), 303–311. 10.1016/j.chembiol.2005.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda M.; Tanaka K.; Hoshino K.; Akiyama H.; Tanimura A.; Saito Y. (1997) Profiles of Potentially Antiallergic Flavonoids in 27 Kinds of Health Tea and Green Tea Infusions. J. Agric. Food Chem. 45 (7), 2561–2564. 10.1021/jf970024y. [DOI] [Google Scholar]
- Yu M. S.; Lee J.; Lee J. M.; Kim Y.; Chin Y.-W.; Jee J.-G.; Keum Y.-S.; Jeong Y.-J. (2012) Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg. Med. Chem. Lett. 22 (12), 4049–4054. 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adedeji A. O.; Singh K.; Calcaterra N. E.; DeDiego M. L.; Enjuanes L.; Weiss S.; Sarafianos S. G. (2012) Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob. Agents Chemother. 56 (9), 4718–28. 10.1128/AAC.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafianos S. G., and Adedeji A. O. (2013) Suppression of SARS replication by SARS helicase inhibitors, WO2013188887A1, World Intellectual Property Organization.
- Kesel A. J. (2006) The bananins: new anticorona-RNA-viral agents with unique structural signature. Anti-Infect. Agents Med. Chem. 5 (2), 161–174. 10.2174/187152106776359039. [DOI] [Google Scholar]
- Narayanan R., Miller D. D., Ponnusamy T., Hwang D.-J., Pagadala J., Duke C. B., Coss C. C., Dalton J. T., and He Y. (2017) Select androgen receptor degrader (SARD) ligands and methods of use thereof, US20170029370A1 and US10017471B2, US Patent and Trademark Office, Washington, DC.
- Lucas J. M.; Heinlein C.; Kim T.; Hernandez S. A.; Malik M. S.; True L. D.; Morrissey C.; Corey E.; Montgomery B.; Mostaghel E.; Clegg N.; Coleman I.; Brown C. M.; Schneider E. L.; Craik C.; Simon J. A.; Bedalov A.; Nelson P. S. (2014) The Androgen-Regulated Protease TMPRSS2 Activates a Proteolytic Cascade Involving Components of the Tor Microenvironment and Promotes Prostate Cancer Metastasis. Cancer Discovery 4 (11), 1310–1325. 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman M. (2020) Sex hormones signal why virus hits men harder. Science (Washington, DC, U. S.) 368 (6495), 1038–1039. 10.1126/science.368.6495.1038. [DOI] [PubMed] [Google Scholar]
- Ghazizadeh Z., Majd S., Richter M., Samuel R., Zekavat S. M., Asgharian H., Farahvashi S., Kalantari A., Ramirez J., and Zhao H.. et al. (2020) Androgen Regulates SARS-CoV Receptor Levels and Is Associated with Severe COVID-19 Symptoms in Men. bioRxiv, DOI: 10.1101/2020.05.12.091082. [DOI] [PMC free article] [PubMed]
- Pomothy J.; Szombath G.; Rokonál P.; Mátis G.; Neogrády Z.; Steinmetzer T.; Pászti -Gere E. (2016) The Impact of Acute Matriptase Inhibition in Hepatic Inflammatory Model. BioMed Res. Int. 2016, 1. 10.1155/2016/6306984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sielaff F.; Böttcher-Friebertshäuser E.; Meyer D.; Saupe S. M.; Volk I. M.; Garten W.; Steinmetzer T. (2011) Development of substrate analogue inhibitors for the human airway trypsin-like protease HAT. Bioorg. Med. Chem. Lett. 21 (16), 4860–4864. 10.1016/j.bmcl.2011.06.033. [DOI] [PubMed] [Google Scholar]
- Steinmetzer T., Meyer D., Hammami M., Sielaff F., Garten W., and Boettcher-Friebertshaeuser E. (2013) Use of TMPRSS2 inhibitors as medicaments, WO2013014074A1, World Intellectual Property Organization.
- Steinmetzer T., Sielaff F., Garten W., Boettcher E., Freuer C., Matrosovich M., and Matrosovich T. (2010) Use of HAT inhibitors and TMPRSS2 inhibitors as medicaments, WO2010149459A1, World Intellectual Property Organization.
- Hardes K.; Ivanova T.; Thaa B.; McInerney G. M.; Klokk T. I.; Sandvig K.; Kuenzel S.; Lindberg I.; Steinmetzer T. (2017) Elongated and Shortened Peptidomimetic Inhibitors of the Proprotein Convertase Furin. ChemMedChem 12 (8), 613–620. 10.1002/cmdc.201700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Pohlmann S. (2020) A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 78 (21), 779–784. 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B.; Valle C.; de Lamballerie X.; Canard B.; Seidah N. G.; Decroly E. (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 176, 104742. 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture F.; Kwiatkowska A.; Dory Y. L.; Day R. (2015) Therapeutic uses of furin and its inhibitors: a patent review. Expert Opin. Ther. Pat. 25 (4), 379–396. 10.1517/13543776.2014.1000303. [DOI] [PubMed] [Google Scholar]
- Imran M.; Saleemi M. K.; Chen Z.; Wang X.; Zhou D.; Li Y.; Zhao Z.; Zheng B.; Li Q.; Cao S.; Ye J. (2019) Decanoyl-Arg-Val-Lys-Arg-Chloromethylketone: An Antiviral Compound That Acts against Flaviviruses through the Inhibition of Furin-Mediated prM Cleavage. Viruses 11 (1011), 1011. 10.3390/v11111011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao G.-S.; Cregar L.; Wang J.; Millis S. Z.; Tang C.; O’Malley S.; Johnson A. T.; Sareth S.; Larson J.; Thomas G. (2006) Synthetic small molecule furin inhibitors derived from 2,5-dideoxystreptamine. Proc. Natl. Acad. Sci. U. S. A. 103 (52), 19707–19712. 10.1073/pnas.0606555104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.; Guo Q.; Zhao L. (2016) Overview of Oroxylin A: A Promising Flavonoid Compound. Phytother. Res. 30, 1765–1774. 10.1002/ptr.5694. [DOI] [PubMed] [Google Scholar]
- Bie B.; Sun J.; Guo Y.; Li J.; Jiang W.; Yang J.; Huang C.; Li Z. (2017) Baicalein: A review of its anti-cancer effects and mechanisms in Hepatocellular Carcinoma. Biomed. Pharmacother. 93, 1285–1291. 10.1016/j.biopha.2017.07.068. [DOI] [PubMed] [Google Scholar]
- Lalou C.; Basak A.; Mishra P.; Mohanta B. C.; Banik R.; Dinda B.; Khatib A. M. (2013) Inhibition of tumor cells proliferation and migration by the flavonoid furin inhibitor isolated from Oroxylum indicum. Curr. Med. Chem. 20 (4), 583–591. 10.2174/0929867311320040010. [DOI] [PubMed] [Google Scholar]
- Majumdar S.; Mohanta B. C.; Chowdhury D. R.; Banik R.; Dinda B.; Basak A. (2010) Proprotein convertase inhibitory activities of flavonoids isolated from Oroxylum indicum. Curr. Med. Chem. 17 (19), 2049–2058. 10.2174/092986710791233643. [DOI] [PubMed] [Google Scholar]
- Worachartcheewan A.; Nantasenamat C.; Naenna T.; Isarankura-Na-Ayudhya C.; Prachayasittikul V. (2009) Modeling the activity of furin inhibitors using artificial neural network. Eur. J. Med. Chem. 44 (4), 1664–1673. 10.1016/j.ejmech.2008.09.028. [DOI] [PubMed] [Google Scholar]
- Becker G. L.; Hardes K.; Steinmetzer T. (2011) New substrate analogue furin inhibitors derived from 4-amidinobenzylamide. Bioorg. Med. Chem. Lett. 21 (16), 4695–4697. 10.1016/j.bmcl.2011.06.091. [DOI] [PubMed] [Google Scholar]
- Shiryaev S. A.; Ratnikov B. I.; Chekanov A. V.; Sikora S.; Rozanov D. V.; Godzik A.; Wang J.; Smith J. W.; Huang Z.; Lindberg I.; Samuel M. A.; Diamond M. S.; Strongin A. Y. (2006) Cleavage targets and the D-arginine-based inhibitors of the West Nile virus NS3 processing proteinase. Biochem. J. 393 (2), 503–511. 10.1042/BJ20051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strongin A., Lebl M., and Day R. (2009) Targeting host proteinases as a therapeutic strategy against viral and bacterial pathogens, US8053553B2, U.S. Patent and Trademark Office, Washington, DC.
- Zhang J., Ma X., Yu F., Liu J., Zou F., Pan T., and Zhang H. (2020) Teicoplanin potently blocks the cell entry of 2019-nCoV. bioRxiv, DOI: 10.1101/2020.02.05.935387.
- Matsuyama S., Kawase M., Nao N., Shirato K., Ujike M., Kamitani W., Shimojima M., and Fukushi S. (2020) The inhaled corticosteroid ciclesonide blocks coronavirus RNA replication by targeting viral NSP15. bioRxiv, DOI: 10.1101/2020.03.11.987016. [DOI] [PMC free article] [PubMed]
- Kang Y.-L., Chou Y.-Y., Rothlauf P. W., Liu Z., Soh T. K., Cureton D., Case J. B., Chen R. E., Diamond M. S., and Whelan S. P. J.. et al. (2020) Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. bioRxiv, DOI: 10.1101/2020.04.21.053058. [DOI] [PMC free article] [PubMed]
- Ohashi H., Watashi K., Saso W., Shionoya K., Iwanami S., Hirokawa T., Shirai T., Kanaya S., Ito Y., and Kim K. S.. et al. (2020) Multidrug treatment with nelfinavir and cepharanthine against COVID-19. bioRxiv, DOI: 10.1101/2020.04.14.039925.
- Yuan X., Yi W., Liu B., Tian S., Cao F., Wang R., Qi B., Lu F., Fang M., and Pei F.. et al. (2020) Pulmonary radiological change of COVID-19 patients with 99mTc-MDP treatment. medRxiv, DOI: 10.1101/2020.04.07.20054767.
- Earhart A. P.; Holliday Z. M.; Hofmann H. V.; Schrum A. G. (2020) Consideration of dornase alfa for the treatment of severe COVID-19 acute respiratory distress syndrome. New Microbes New Infect. 35, 100689. 10.1016/j.nmni.2020.100689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakom S.; Iorio F.; Eyers P. A.; Escott K. J.; Hopper S.; Wells A.; Doig A.; Guilliams T.; Latimer J.; McNamee C.; Norris A.; Sanseau P.; Cavalla D.; Pirmohamed M. (2019) Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discovery 18, 41–58. 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- Seyhan A. A. (2019) Lost in translation: the valley of death across preclinical and clinical divide - identification of problems and overcoming obstacles. Trans. Med. Commun. 4, 18. 10.1186/s41231-019-0050-7. [DOI] [Google Scholar]
- Cavasotto C., and Di Filippo J. (2020) In silico Drug Repurposing for COVID-19: Targeting SARS-CoV-2 Proteins through Docking and Quantum Mechanical Scoring. ChemRxiv, DOI: 10.26434/chemrxiv.12110199.v2. [DOI] [PubMed]
- Martin W. R., and Cheng F. (2020) Repurposing of FDA-Approved Toremifene to Treat COVID-19 by Blocking the Spike Glycoprotein and NSP14 of SARS-CoV-2. ChemRxiv, DOI: 10.26434/chemrxiv.12431966. [DOI] [PMC free article] [PubMed]
- Zhou Y., Hou Y., Shen J., Kallianpur A., Zein J., Culver D. A., Farha S., Comhair S., Fiocchi C., and Gack M. U.. et al. (2020) A Network Medicine Approach to Investigation and Population-based Validation of Disease Manifestations and Drug Repurposing for COVID-19. ChemRxiv, DOI: 10.26434/chemrxiv.12579137. [DOI] [PMC free article] [PubMed]
- Zeng X., Song X., Ma T., Pan X., Zhou Y., Hou Y., Zhang Z., Li K., Karypis G., and Cheng F. (2020) Repurpose Open Data to Discover Therapeutics for COVID-19 using Deep Learning. J. Proteome Res. 10.1021/acs.jproteome.0c00316. [DOI] [PubMed] [Google Scholar]
- Elmezayen A. D.; Al-Obaidi A.; Sahin A. T.; Yelekci K. (2020) Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. 1. 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowski T., Chen L., Eastman R., Itkin Z., Shinn P., Chen C., Guo H., Zheng W., Michael S., and Simeonov A.. et al. (2020) Discovery of Synergistic and Antagonistic Drug Combinations against SARS-CoV-2 In Vitro. bioRxiv, DOI: 10.1101/2020.06.29.178889. [DOI] [PMC free article] [PubMed]
- Zhou Y.; Hou Y.; Shen J.; Huang Y.; Martin W.; Cheng F. (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery 6 (1), 14. 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.