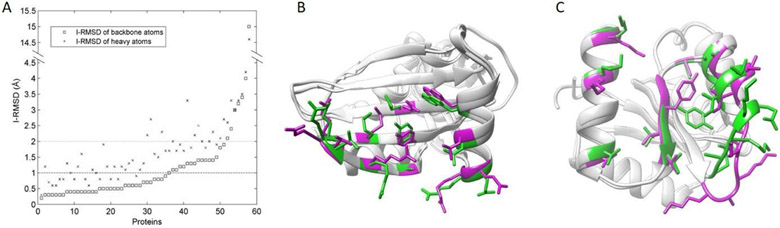
Figure 3.
(A) I-RMSDs of bound and unbound protein structures. Both the backbone I-RMSD and the heavy atom I-RMSD are plotted. Proteins are sorted by values of the backbone I-RMSD. (B) and (C) show two examples of interface residues in bound (green) and unbound (magenta) protein structures. The proteins are matched by the UCSF Chimera version 1.11 [36]. (B) Values of I-RMSD_b and I-RMSD_h between bound (PDB entry: 2g30, chain A) and unbound (PDB entry: 2iv9, chain A) protein structures in (B) are 0.5 Å and 1.9 Å, respectively. (C) Values of I-RMSD_b and I-RMSD_h between bound (PDB entry: 2b1j, chain A) and unbound (PDB entry: 3rvq, chain A) protein structures in (B) are 2.4 Å and 3.3 Å, respectively.