Figure 1.
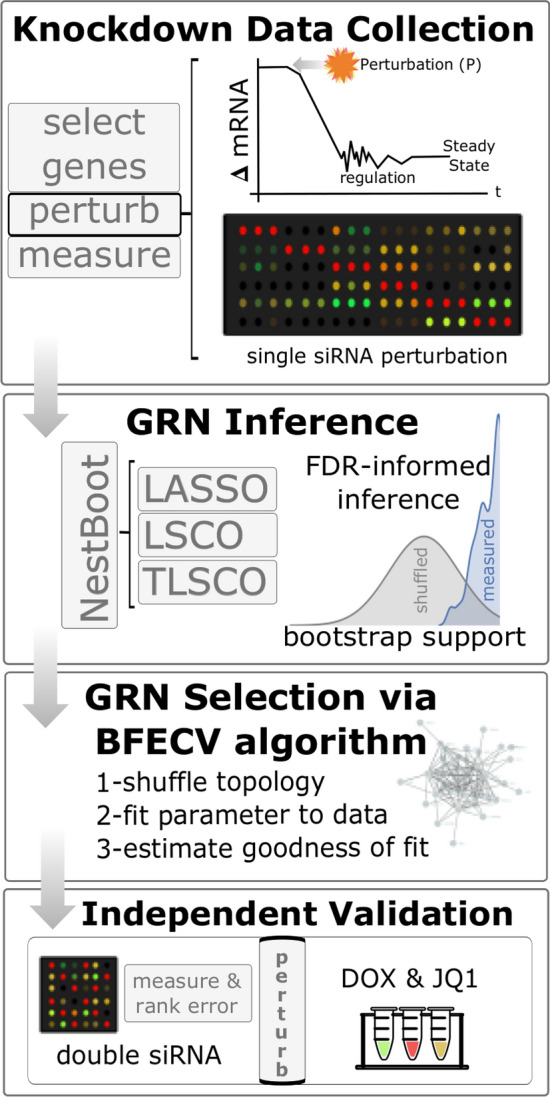
Workflow of project. siRNA perturbation experiments were carried out systematically per gene, resulting in a change in mRNA level which elicits a regulatory response over time before reaching steady state when gene expression was measured. GRN inference: nested bootstrapping was applied to three inference methods, producing GRNs with an FDR set to 5%. GRN Selection: Estimate goodness of fit with the Balanced Fitting of Errors framework under cross-validation (BFECV) and compare to shuffled topologies (Algorithm 1). Independent Validation: Each inferred GRNs’ ability to predict an independent dataset was evaluated in comparison to a distribution of shuffled topologies. Finally, the overall most predictive GRN was selected. Two novel links were experimentally validated.
