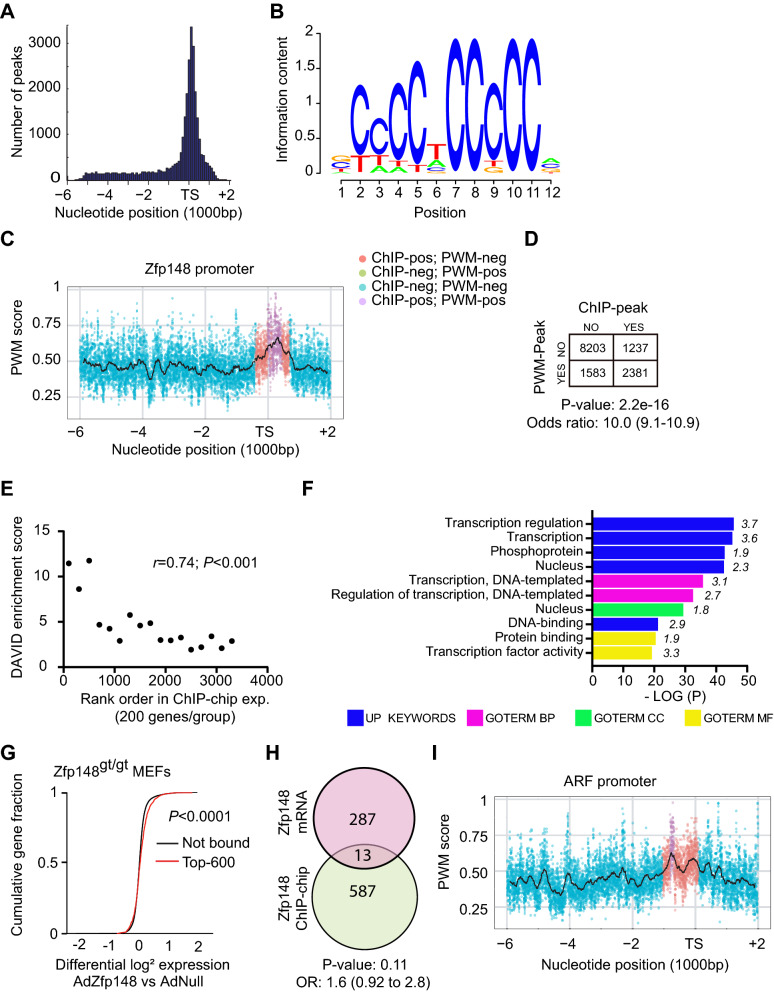
Figure 3.
Zfp148 binds to clustered cytosine-rich elements in promoters of other transcription factors. (A) Frequency plot showing the location of binding sites of AdZfp148FLAG in relation to transcription starts (TS) for one representative ChIP-chip experiment. (B) Position Weight Matrix (PWM)-logo for Zfp148. (C) A representative gene (Zfp148) with both ChIP-peaks and PWM-peaks. Each point represents the PWM score at the specific position, taking only the best score of the two strands. The black line indicates the mean score over the current position in a 200 bp surrounding window. The intersect of ChIP- and PWM-peaks is indicated by a color code (explained in the figure). (D) Contingency table showing the distribution of promoters with or without ChIP- and/or CWM-peaks. 95% confidence interval in brackets. (E) Graph showing maximum DAVID enrichment score on y-axis and rank order on x-axis for bins of 200 genes based on ChIP-binding score. r indicates Spearman correlation. (F) Graph showing enriched functional terms for the top-600 AdZfp148FLAG binding genes. Fold-enrichment is shown in italics. − LOG10 P-value, negative logarithm with base 10 of the adjusted p-value; UP, UniPRot; GOTERM, gene ontology term; CC, cell compartment; MF, molecular function; BP, biological process. (G) Cumulative distribution function (CDF) of log2-transformed gene expression ratios in Zfp148gt/gt MEFs transduced with AdZfp148 versus AdNull. Red line, genes that bind AdZfp148FLAG with high affinity (n = 600 based on rank order); black line, genes that did not bind AdZfp148FLAG in the ChIP-chip experiments (n = 10,163) (expression analysis, n = 3; ChIP-chip analysis, n = 4). (H) Venn diagram showing the intersect of the top-300 genes that were downregulated in Fig. 1 (red) and the top-600 AdZfp148FLAG binding genes (green). 95% confidence interval in brackets. (I) Graph showing ChIP scores and PWM scores for the ARF promoter. Data representation as in (C).