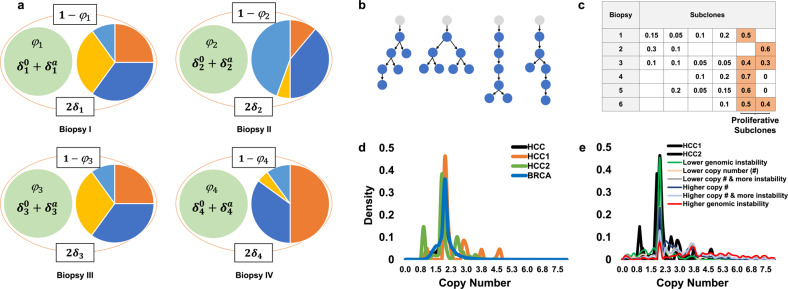
Fig. 2. Our model for the effects of copy number alterations on mutated-read fractions in DNA profiles.
a For each mutation in each biopsy s, the mutated-read fraction is a function of the true proportion of profiled cells with the mutation (mutation frequency) φs, the copy number of the reference allele in cells without the mutation δs, and the copy numbers of the reference and the mutated allele in tumor cells with the mutation, and , respectively. The frequency of tumor and WT cells without the mutation is (1 − φs) and the two reference alleles can be assumed to have a combined copy number of 2δs. c–e To compare inference methods we synthetically generated profiling data based on parametrized simulated copy number distributions. b Shown are representative phylogenies and c a representative cellular composition matrix, as well as d density plots of average copy numbers across profiles of TCGA-profiled hepatocellular (HCC) and breast (BRCA) carcinomas. These include the distribution of copy numbers in two individual HCCs (HCC1 and HCC2) and across all HCCs (black) and all BRCAs (blue); copy numbers ranged from 0 to >260×. e Simulated copy numbers, including simulations of more-stable and less-stable cancers, ranged from 0× to 15× copies.