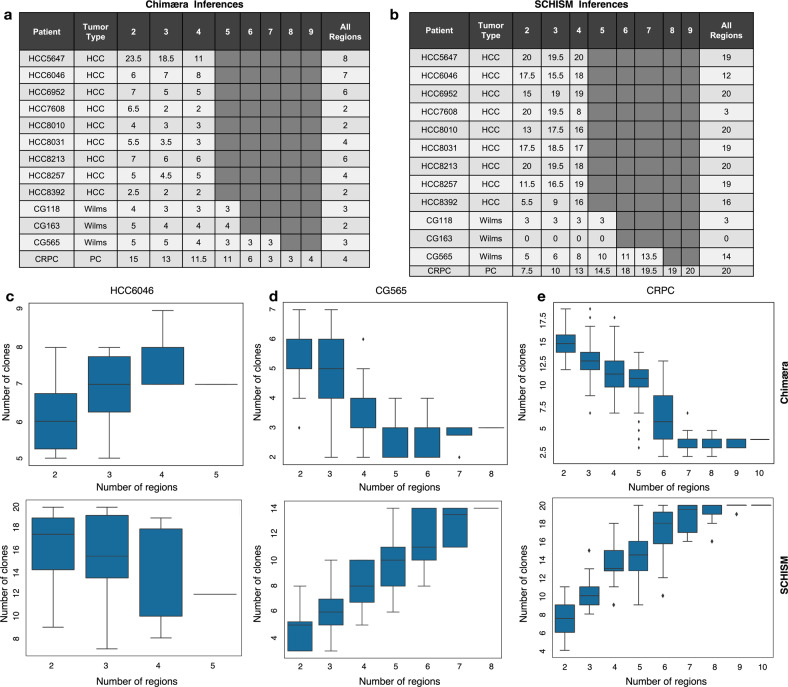
Fig. 4. The number of predicted subclones depends on the number of profiled regions per tumor.
a, b Lin et al. profiled five regions of each of nine HBV-positive HCCs. We profiled 6–8 regions of each of three Wilms’ tumors and ten regions of a castrate-resistant prostate cancer (CRPC). For each tumor, we exhaustively selected all region subsets size-2 and up, and compared the number of predicted tumor subclones across subsets to those obtained using all available region profiles using a Chimæra and b SCHISM. Chimæra analysis of any 4-tumor regions resulted in a similar number of subclones as analysis of five regions, however, profiling two regions produced fewer predicted subclones. SCHISM performed better on stable genomes (CG118) than on unstable genomes (CG163 and CRPC). c Chimæra analyses (top) of HCC6046 profiles suggested convergence of subclone predictions using three profiled regions, while SCHISM analyses (bottom) produced a higher prediction variability across profiling subsets. d Similarly, Chimæra analyses of four regions of CG565 produced a similar number of clones as profiles of eight regions. e Chimæra analyses of seven regions of our CRPC tumor produced a similar number of clones as analysis of ten regions; however, analyses based on five or fewer tumor areas produced significantly more predicted tumor subclones due to reduced aggregating power. SCHISM analyses based on six or more regions produced consistent counts of predicted subclones, but this number was considerably greater than the number of subclones predicted by Chimæra.