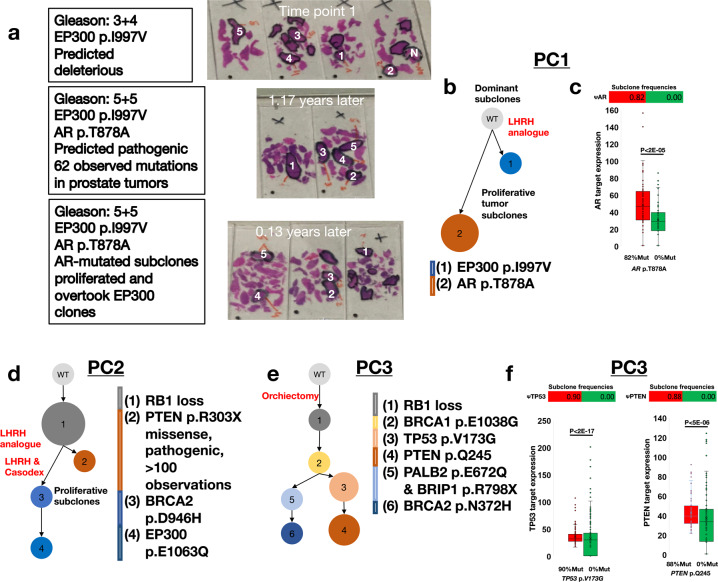
Fig. 7. Inferred phylogenies of tumors from three prostate cancer patients.
a Five regions of Prostate Cancer 1 (PC1) were profiled at each of three time points, identifying potentially deleterious mutations at each time point. b PC1’s inferred phylogeny suggested that EP300 p.I997V mutation was present at Time Point 1, and that the tumor subclone with EP300 p.I997V (Subclone 1) is distinct from the subclone with AR p.T878A (Subclone 2), which was observed only in the later time point and whose clonal frequency increased with time. c Differential protein expression analysis suggested that regions that were predicted to have high clonal composition of Subclone 2 (82% vs. 0%) had higher AR-target expression than regions without Subclone 2. d Inferred phylogeny for Prostate Cancer 2 (PC2) suggested that RB1 loss was an initiating event, and that that majority of tumor cells are the results of divergent evolution following the acquisition of PTEN and BRCA2 mutations, Subclones 2 and 3, respectively. In total, 5 regions of PC2 were profiled in each of the five Time Points; Subclone 1 was observed in all time points, and Subclones 3 and 4 in Time Points 4 and 5. e PC3’s inferred phylogeny included RB1 loss as an initiating event, followed by the acquisition of a BRCA1 mutation. These tumor cells then either acquired TP53 and PTEN mutations, or PALB2, BRIP1, and BRCA2 mutations. All mutations and subclones were observed in each of the two time points profiled. The PTEN, BRCA1, and BRCA2 mutations were previously observed in cancers. f A comparison of PC3 regions with low and high composition for Subclones 3 and 4, 90% vs. 0% and 88% vs. 0%, respectively, suggested significant differential protein expression for targets of TP53 and PTEN.