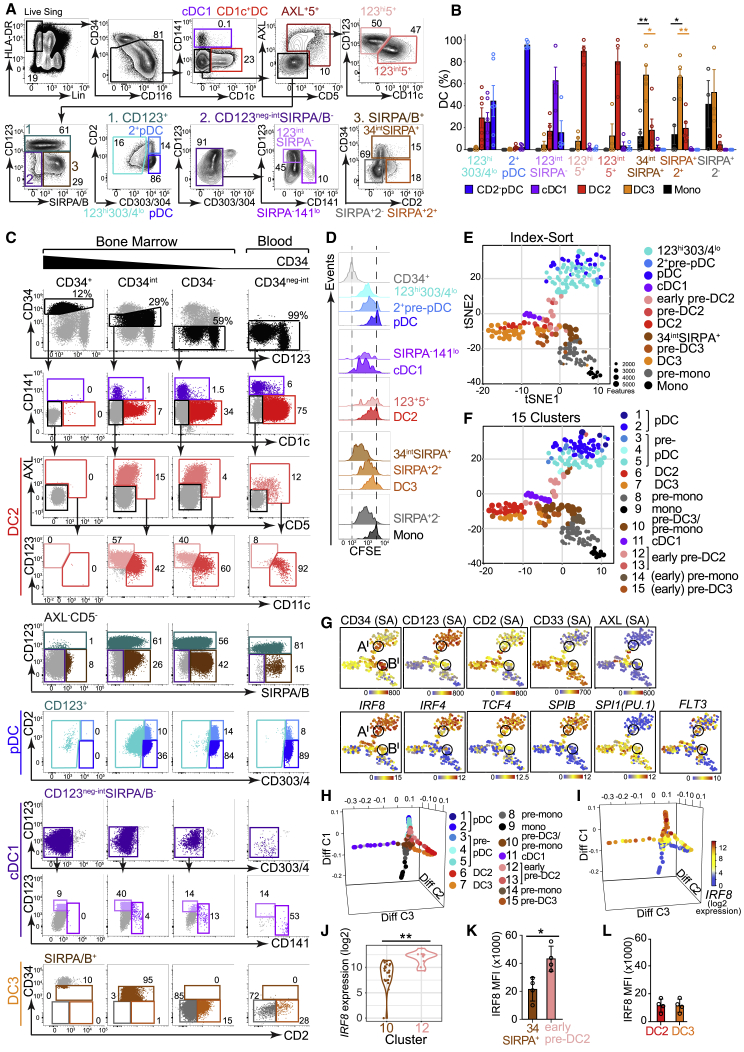
Figure 4.
Two Trajectories of DC Development Connect the Progenitor Compartment with Mature DCs
(A) Flow gating strategy used to identify DCs and their precursors in BM, including CD141+ cDC1s; CD1c+ DCs; AXL+CD5+ cells composed of CD123hiCD11c− (light pink) and CD123intCD11c+ (dark pink) fractions; CD2+ (light blue) and CD2− (dark blue) pDCs; CD123+CD303/4lo cells (turquoise); SIRPA/B−CD123intCD141− (lightest purple) and CD141lo (light purple) populations; and CD123−SIRPA/B CD34int (brown), CD34−CD2+ (dark orange), and CD34−CD2− (gray) precursors.
(B) The output of in vitro culture of CD34int DC precursors FACS-purified from BM using the gating strategy described in (A). Population-specific output is expressed as a proportion (%) of the total cells captured by all DC and monocyte gates. CD123hi303/4lo (six experiments from four donors; 6;4); CD2+ pre-pDCs (5;3); CD123hi5+ (4;3); CD123int5+ (4;3); CD34intCD123int (4;4); CD34intSIRPA+ (5;5); SIRPA+2+ (4;4); SIRPA+2− (4;4). Bars represent mean + SEM, and circles represent individual experiments. Significant differences in the proportional output of DC2s versus DC3s (red) or DC3s versus monocyte (black) are indicated: ∗p < 0.05; ∗∗p < 0.01 (paired, two-tailed t test).
(C) Flow gating strategy from (A) applied to lin−HLA-DR+ cells from HC BM fractionated by high, intermediate, and low CD34 expression, next to blood (columns) for comparison of antigen expression levels among progenitor, precursor, and mature populations. Individual DC lineages are ordered in rows.
(D) Proliferative potential of FACS-purified DC and DC precursors estimated by CFSE dilution (see STAR Methods). CD34+progenitors and CD14+monocytes were included as positive and negative controls, respectively. The CFSE dilution histograms for each precursor are grouped and ordered according to their proposed position in the developmental trajectory for each DC lineage. Plots shown are representative of n = 3 experiments (summarized in Figure S4H).
(E and F) tSNE visualization of the first 20 principal components (explaining 35% total variance) of the transcriptomes of 244 single cells adaptively sampled from lin−HLA-DR+ CD34neg-int precursor and mature DC populations of BM. tSNE plots are annotated by the gate of origin from index-linked flow (E) or by 15 clusters generated from hierarchical clustering of all protein-coding non-cell-cycle genes (F), independently of surface phenotype (Figure S4K).
(G) Heatmaps showing the expression of key surface antigens (SAs) (index-linked flow) or log2 gene expression of TFs and FLT3 (scRNA-seq) across the tSNE plot in (E) and (F). Black circles represent regions of high or low IRF8 expression, marked Al or Bl, respectively. The differential expression patterns of these regions correspond to the patterns of regions “A” (IRF8hiCD123intGMP) and “B” (IRF8loGMP33+) in Figures 3E–3G.
(H and I) Diffusion map generated with all protein-coding, non-cell-cycle genes to infer pseudo-temporal ordering of cells and reconstruct lineage branching. (H) Cells are colored according to the hierarchical clusters generated in Figure S4K. (I) IRF8 expression (log2). Diff C, diffusion component.
(J) Violin plot of differential IRF8 expression (log2) in clusters 10 (SIRPA+34int) and 12 (early pre-DC2). ∗∗p < 0.001 by Mann-Whitney U.
(K and L) MFI of intracellular IRF8 by flow analysis across gates identifying BM 34intSIRPA+ pre-DC3s and pre-mono and CD123hiCD5+ early pre-DC2s (K) and CD5+ DC2s and CD5− DC3s (L) (n = 4). ∗p = 0.028 by Mann-Whitney U.
See also Figure S4.