Figure 4.
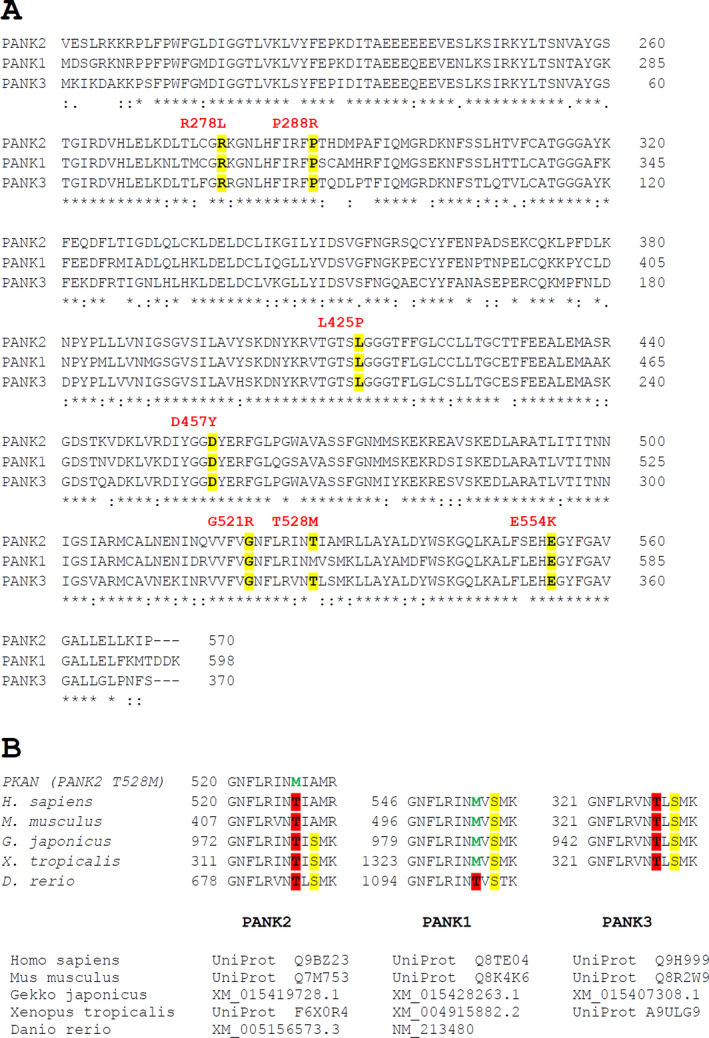
Conservation of PANK2 point mutations sites between the isoforms and phylogenetic variation within a partial sequence corresponding to the region around T528 of human PANK2. (A) Sequence alignment of the three human PANK isoforms was performed with the CLUSTALW algorithm using the Uniprot sequences of human PANK2 (Q9BZ23), PANK1 (Q8TE04), and PANK3 (Q9H999) with * (asterisk) and: (colon) indicating full conservation and conservation between groups of amino acids with highly similar properties, respectively. The seven PANK2 point mutations relevant for this study are indicated above the respective positions in red and amino acid conservation is highlighted in yellow. (B) Phylogenetic comparisons of PANK2 sequences around T528 between two mammalian, one reptile, one amphibian, and one fish species (species names and sequence identifiers below the alignments) are shown for the three PANK isoforms as indicated. Please note that the respective threonine corresponding to T528 of human PANK2 (highlighted in red) is conserved in PANK2 and PANK3 isoforms while it shifted to methionine (green) early in the evolution of the PANK1 protein. Conversely, the alanine at position 530 of human PANK2 is only found in mammalian PANK2 isoforms while phylogenetically distant species have a serine at this position (highlighted in yellow), which is also conserved in PANK1 and PANK3 isoforms.
