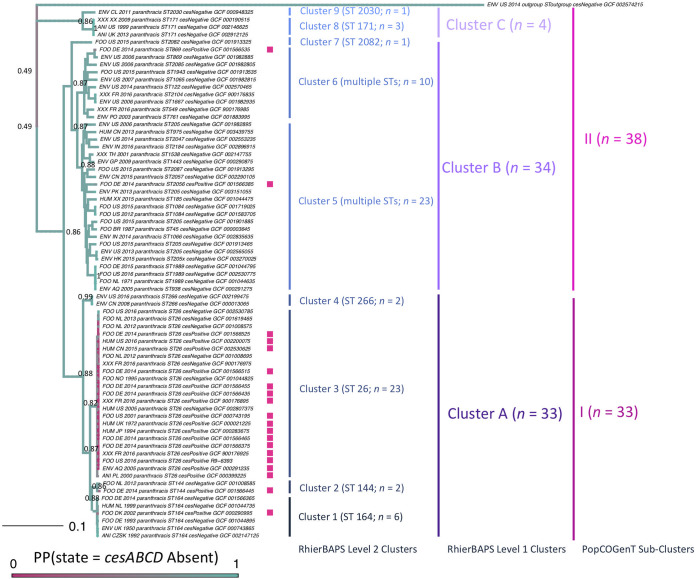
FIG 1.
Maximum-likelihood phylogeny constructed using core SNPs identified among 71 emetic group III B. cereus sensu lato genomes and their closely related, nonemetic counterparts, plus outgroup genome B. cereus sensu lato strain AFS057383. Tip labels of genomes that possess cereulide synthetase-encoding genes cesABCD are annotated by a pink square. Clade labels correspond to (i) RhierBAPs level 2 cluster assignments, denoted as clusters 1 to 9, with the number of isolates assigned to a cluster (n) and sequence type (ST) determined using in silico multilocus sequence typing (MLST) listed in parentheses; (ii) RhierBAPs level 1 cluster assignments, denoted as clusters A, B, and C; (iii) PopCOGenT subcluster assignments, denoted as I and II. Tree edge and node colors correspond to the posterior probability (PP) of being in a ces-negative state, obtained using an empirical Bayes approach, in which a continuous-time reversible Markov model was fitted, followed by 1,000 simulations of stochastic character histories using the fitted model and tree tip states. Equal root node prior probabilities for ces-positive and ces-negative states were used. Node labels denote selected PP values, chosen for readability. The phylogeny was rooted along the outgroup genome, and branch lengths are reported in substitutions/site.