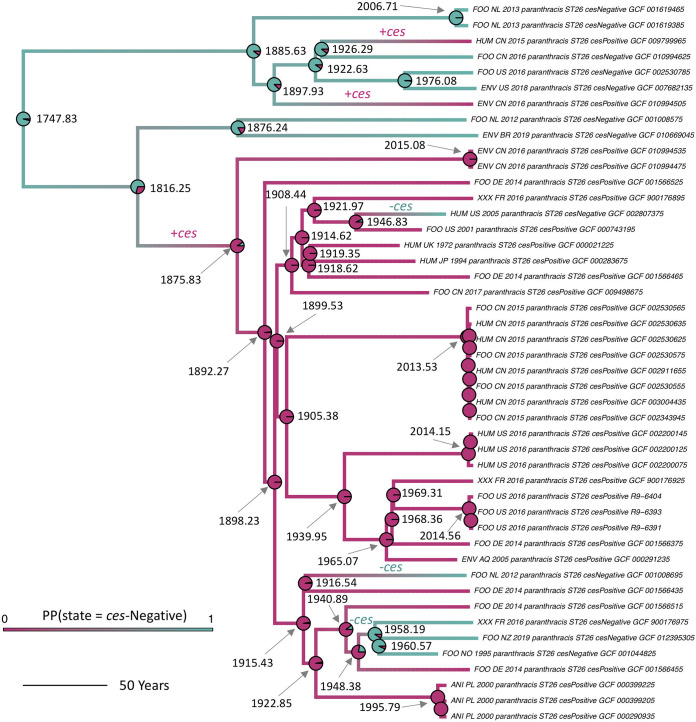
FIG 4.
Rooted, time-scaled maximum clade credibility (MCC) phylogeny constructed using core SNPs identified among 46 group III B. cereus sensu lato genomes assigned to sequence type (ST) 26. Branch color corresponds to posterior density, denoting the probability of a lineage being in a ces-negative state as determined using ancestral state reconstruction. Pie charts at nodes denote the posterior probability (PP) of a node being in a ces-negative (teal) or ces-positive (pink) state. Labels along branches denote a ces gain or loss event (denoted by +ces or –ces, respectively). Node labels indicate node ages in years (selected for readability) and are either placed to the right of their corresponding node pie chart or connected to their corresponding node pie chart with a gray arrow. Branch lengths are reported in years. Core SNPs were identified using Snippy v4.3.6. The phylogeny was constructed using the results of five independent runs using a relaxed lognormal clock model, the Standard_TVMef nucleotide substitution model, and the Coalescent Bayesian Skyline population model implemented in BEAST v2.5.1, with 10% burn-in applied to each run. LogCombiner-2 was used to combine BEAST 2 log files, and TreeAnnotator-2 was used to construct the phylogeny using median node heights. Ancestral state reconstruction was performed using a prior on the root node in which the probability of the ST 26 ancestor being ces positive or ces negative was 0.2 and 0.8, respectively (see Tables S2 and S5 in the supplemental material). For ST 26 ancestral state reconstruction results obtained using different root node priors and isolate sets, see Table S5 and see Fig. S11 to S18 (https://doi.org/10.6084/m9.figshare.c.5057276.v1).