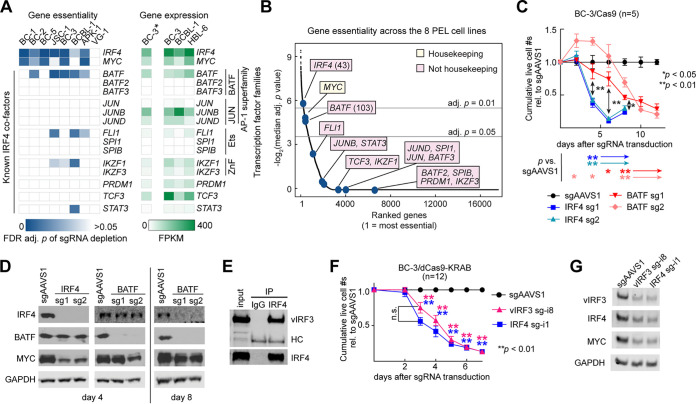
FIG 1.
Cellular BATF and KSHV vIRF3 are candidates for essential cofactors and regulators of IRF4. (A to D) BATF, a cellular co-TF of IRF4 in other settings, is essential across PEL cell lines. (A) (Left) Blue heat map showing essentiality in eight PEL cell lines of genes that are cofactors of IRF4 in other settings. The heat map depicts false-discovery-rate (FDR)-adj. P values of depletion of the sgRNAs targeting these genes in our recent PEL CRISPR/Cas9 gene essentiality screens (21). VG-1 cells performed relatively poorly in these screens, but we have previously shown that IRF4 is similarly essential in VG-1 (36). While BATF did not meet our essentiality cutoff (for example, in BC-1) in our 14-day screens, BATF essentiality in BC-1 was confirmed (see Fig. S3). (Right) Green heat map showing expression (in fragments per kilobase [of transcript] per million mapped reads, FPKM) of the same genes in a BC3 RNA-Seq data set generated in this study (marked by an asterisk; see below) and published RNA-Seq data (42). (B) All genes screened previously by Manzano et al. (21) were ranked by their median FDR-adj. P values of sgRNA depletion (negative log2-transformed) across the 8 PEL cell lines. Both IRF4 and BATF scored among the most essential genes across the eight PEL cell lines. Yellow indicates that MYC scored as essential in all 16 cancer cell types analyzed in this study and is therefore a “housekeeping gene,” while pink boxes denote genes that are less commonly essential in different types of cancer. (C) CRISPR/Cas9-mediated KO of IRF4 or BATF in PEL cell line BC-3 showed that each triggered a strong, but temporally distinct, decrease in live-cell numbers over time, relative to the negative-control sgAAVS1, which is directed to an intergenic safe harbor locus. BC-3/Cas9 cells were transduced with lentiviruses expressing two individual sgRNAs targeting the indicated genes, each delivered at an MOI of 1. See Fig. S2 and S3 for data from three additional PEL cell lines. (D) Western blot analyses of the expression of IRF4, BATF, the IRF4 downstream target MYC, and the loading control GAPDH (glyceraldehyde-3-phosphate dehydrogenase) on day 4 or day 8 following CRISPR/Cas9-mediated KO of IRF4 or BATF as shown in panel C. Results suggest that BATF positively regulates IRF4 expression. See Fig. S1 for quantification across replicates and Fig. S2 and S3 for data from three additional PEL cell lines. (E) Immunoprecipitation of endogenous IRF4 from partially DNase-digested BC-3 nuclear extracts efficiently coprecipitates endogenous KSHV vIRF3, representative of n = 3. Quantification of Li-Cor Western results showed that between ∼15% and ∼49% of input vIRF3 coprecipitated with IRF4. See Fig. S4 for reciprocal coimmunoprecipitation of ectopically expressed proteins. HC, heavy chain. (F) CRISPRi-mediated repression of vIRF3 or IRF4 expression triggers a temporally similar decrease in live-cell numbers over time. BC-3/dCas9-KRAB cells were transduced with lentiviruses expressing sgRNAs targeting locations near the TSSs of the indicated genes (“sg-i”), each delivered at an MOI of 1. See Fig. S5 for data from the additional PEL cell line BC-1 and Fig. S6 for rescue experiments that confirmed the specificity of this result. Comparable results were obtained with an additional vIRF3-specific sgRNA (sg-i2; see Fig. 4). (G) Western blot analyses of the expression of vIRF3, IRF4, MYC, and the loading control GAPDH on day 4 of experiments performed as described for panel F. See Fig. S5A for quantification over replicates. We note that knockdown of the targeted genes in these analyses was incomplete because the cells underwent cell death before complete knockdown was observed and samples were collected when a substantial portion of live cells remained. Throughout the figure, error bars represent SEM with numbers of biological replicates indicated. *, P < 0.05; **, P < 0.01 (paired two-sided Student's t tests). n.s., not significant.