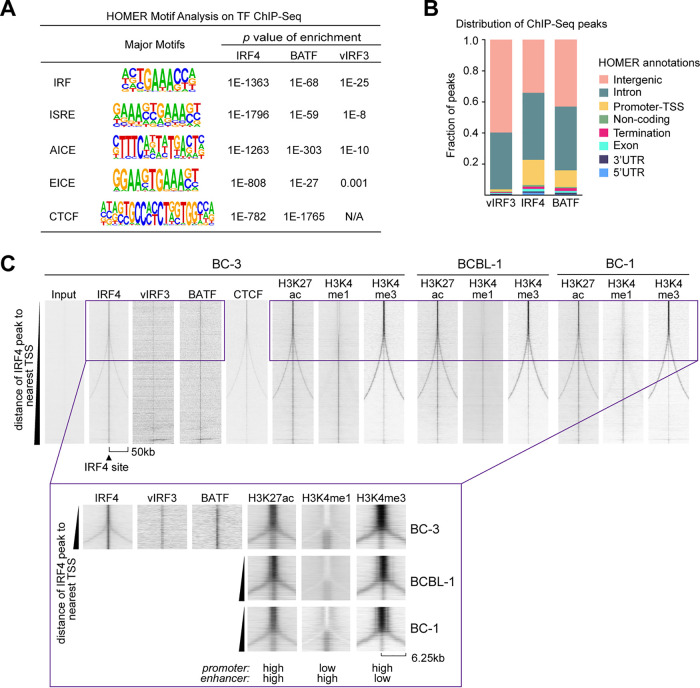
FIG 2.
IRF4, vIRF3, and BATF cooccupy genomic sites. (A) HOMER motif analysis identifies significant enrichment of the IRF motif (i.e., the IRF4 binding site) and the ISRE and AICE motifs in BC-3 IRF4, BATF, and vIRF3 ChIP-Seq data. We also detected enrichments for the composite EICE motif and the CTCF binding site. (B) Distribution of location annotations of ChIP-Seq peaks as determined by HOMER. By default, HOMER considers the −1-kb to +100-bp region of the TSS “Promoter-TSS” and considers the −100-bp to +1-kb region of the transcriptional termination site “Termination.” UTR, untranscribed region. (C) IRF4 ChIP-Seq peaks in BC-3 were sorted by the distance (scale = 50 kb on either end) to the nearest TSS and assessed for cooccupancy with vIRF3, BATF, and CTCF as well as nucleosome modification by H3K27ac, H3K4me1, and H3K4me3 in BC-3, BCBL-1, and BC-1 cells. This analysis was performed using EaSeq (80). The insets at the bottom of the panel show enlarged versions of the boxed portion at the top of the heat map, to highlight differential chromatin modifications at promoters and enhancers. Note that the lack of diverging signal (“fork”) in the vIRF3 or BATF ChIP-Seq compared to the IRF4 ChIP-Seq might be due to the inefficient performance of the antibodies in ChIP experiments.